Q8BI55
Gene name |
Dyrk4 |
Protein name |
Dual specificity tyrosine-phosphorylation-regulated kinase 4 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:101320 |
EC number |
2.7.12.1: Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
Protein Class |
|
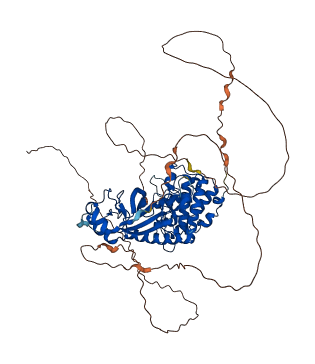
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
364-384 (Activation loop from InterPro)
Target domain |
219-515 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q8BI55
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8BI55-F1 | Predicted | AlphaFoldDB |
28 variants for Q8BI55
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388863270 | 72 | I>F | No | EVA | |
| rs3388856627 | 111 | Q>* | No | EVA | |
| rs3388826843 | 121 | L>V | No | EVA | |
| rs1133908308 | 137 | T>I | No | EVA | |
| rs3388863798 | 193 | E>* | No | EVA | |
| rs3388853406 | 233 | V>G | No | EVA | |
| rs224602281 | 315 | H>Q | No | EVA | |
| rs3388865069 | 329 | I>F | No | EVA | |
| rs3388853408 | 381 | Q>E | No | EVA | |
| rs3388839743 | 381 | Q>R | No | EVA | |
| rs3388861095 | 391 | I>N | No | EVA | |
| rs3388865074 | 396 | Y>* | No | EVA | |
| rs3388865000 | 399 | A>G | No | EVA | |
| rs3388844998 | 408 | I>V | No | EVA | |
| rs3388861389 | 434 | V>L | No | EVA | |
| rs217012827 | 441 | H>D | No | EVA | |
| rs3388860070 | 453 | F>S | No | EVA | |
| rs3388861120 | 454 | D>E | No | EVA | |
| rs3388864662 | 466 | R>* | No | EVA | |
| rs3388844999 | 516 | H>Q | No | EVA | |
| rs3388853341 | 526 | K>N | No | EVA | |
| rs3388868445 | 549 | Q>* | No | EVA | |
| rs3388853339 | 583 | S>N | No | EVA | |
| rs1135293876 | 584 | S>F | No | EVA | |
| rs3388868468 | 587 | H>N | No | EVA | |
| rs3397385889 | 602 | G>V | No | EVA | |
| rs47087308 | 604 | S>P | No | EVA | |
| rs864280737 | 631 | I>V | No | EVA |
No associated diseases with Q8BI55
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.12.1 | Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| peptidyl-threonine phosphorylation | The phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8NE63 | HIPK4 | Homeodomain-interacting protein kinase 4 | Homo sapiens (Human) | PR |
| Q9H422 | HIPK3 | Homeodomain-interacting protein kinase 3 | Homo sapiens (Human) | SS |
| Q92630 | DYRK2 | Dual specificity tyrosine-phosphorylation-regulated kinase 2 | Homo sapiens (Human) | PR |
| Q13627 | DYRK1A | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | Homo sapiens (Human) | PR |
| Q9H2X6 | HIPK2 | Homeodomain-interacting protein kinase 2 | Homo sapiens (Human) | EV |
| Q9NR20 | DYRK4 | Dual specificity tyrosine-phosphorylation-regulated kinase 4 | Homo sapiens (Human) | PR |
| Q61214 | Dyrk1a | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | Mus musculus (Mouse) | PR |
| Q3V016 | Hipk4 | Homeodomain-interacting protein kinase 4 | Mus musculus (Mouse) | PR |
| Q9QZR5 | Hipk2 | Homeodomain-interacting protein kinase 2 | Mus musculus (Mouse) | SS |
| Q4V793 | Hipk4 | Homeodomain-interacting protein kinase 4 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQLLRLPALT | RTETSMDTNK | ARKRSLTTFP | ILKARKKQSF | TSVKVESKPL | GHLQKPSSKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KKLRVNRCPQ | KIPSNTAFPF | VDTKGKRNTV | NFPQIGNKVP | SKGPMQYQEN | QIHNQVLSSE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LKTSEIPFNI | NTKAQDTKPH | PELQKKHKVP | LTVAEALKFF | KNQLSPYEQS | EILGYSELWF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LGLEAKKLNV | VPEKFSKTSF | DDEHGSYMKV | LHDHIAYRYE | VLEMIGKGSF | GQVAKCLDHK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NNELVALKII | RNKKRFHHQA | LVELKILEAL | RRKDKDNNHN | VVHMKDFFYF | RNHLCITFEL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LGINLYELMK | NNSFHGFNLS | IVRRFTFSIL | KCLHMLYVEK | IIHCDLKPEN | IVLYQRGQVT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VKVIDFGSSC | YEHQKVYTYI | QSRFYRSPEV | ILGHPYNMAI | DMWSLGCIMA | ELYTGYPLFP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GENEVEQLAC | IMEVLGLPPA | HFTQTASRRQ | VFFDSKGLPK | NINNNRGGKR | YPDSKDLTMV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VKTYDSSFLD | FLRRCLVWEP | SLRMTPEQAL | KHAWIHEPRK | FKPRPKPQIL | RKPGASISSE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ISTEKAEEQQ | ASKGKKDEAT | KETTDKLKDE | AEKHLENSGK | QQSSVEHTAD | TIQLPHLTEA |
| 610 | 620 | 630 | |||
| SGKSETVAGS | EMSAEQQSTS | SPKSTNILPP | IV |