Q86WR6
Gene name |
C17orf64 |
Protein name |
Uncharacterized protein C17orf64 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:124773 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
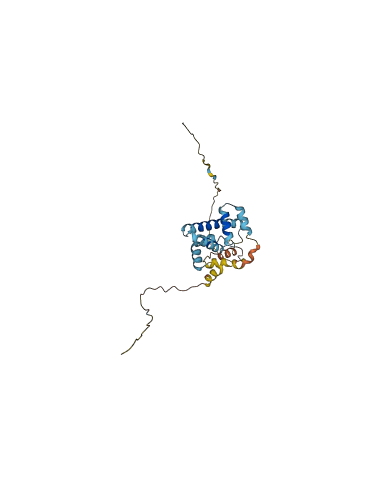
Autoinhibited structure

Activated structure

1 structures for Q86WR6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q86WR6-F1 | Predicted | AlphaFoldDB |
217 variants for Q86WR6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA400773185 rs1191898736 |
2 | E>* | No |
ClinGen TOPMed |
|
|
CA400773207 rs1372711933 |
3 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs753736037 CA8686862 |
5 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA292696829 rs985273637 |
6 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
rs953439254 CA292696828 |
6 | G>W | No |
ClinGen TOPMed |
|
|
CA400773241 rs1432817224 |
7 | Q>E | No |
ClinGen gnomAD |
|
|
rs1362414773 CA400773263 |
8 | G>V | No |
ClinGen gnomAD |
|
|
rs1383704347 CA400773285 |
10 | E>A | No |
ClinGen TOPMed gnomAD |
|
|
rs779607346 CA8686867 |
11 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8686871 rs200863948 CA8686870 |
12 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8686869 rs79813353 |
12 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1369072041 CA400773350 |
15 | L>I | No |
ClinGen TOPMed |
|
|
rs1210050849 CA400773354 |
15 | L>P | No |
ClinGen gnomAD |
|
|
rs1429963533 CA400773381 |
17 | Q>R | No |
ClinGen TOPMed |
|
|
rs1177831117 CA400773814 |
18 | V>M | No |
ClinGen gnomAD |
|
|
rs1164361948 CA400773830 |
19 | T>K | No |
ClinGen TOPMed |
|
|
CA8686884 rs750371587 |
23 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs750371587 CA400773881 |
23 | C>S | No |
ClinGen ExAC gnomAD |
|
|
CA400773884 rs1410979532 |
23 | C>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA292697203 rs1018473051 |
25 | E>V | No |
ClinGen TOPMed |
|
|
CA400773904 rs1327584021 |
26 | T>K | No |
ClinGen gnomAD |
|
|
CA400773908 rs1391790981 |
27 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1435186346 CA400773911 |
27 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400773909 rs1391790981 |
27 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
CA400773920 rs1598333338 |
29 | S>G | No |
ClinGen Ensembl |
|
|
CA8686885 rs372836275 |
29 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA8686887 rs202020451 |
30 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA292697204 rs1030544423 |
30 | A>V | No |
ClinGen Ensembl |
|
|
rs757742869 CA292697205 |
34 | R>K | No |
ClinGen Ensembl |
|
|
rs1222567913 CA400773963 CA400773962 |
35 | D>E | No |
ClinGen gnomAD |
|
|
rs754589179 CA8686888 |
36 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA292697206 rs954849767 |
38 | M>K | No |
ClinGen TOPMed |
|
|
rs954849767 CA400773979 |
38 | M>T | No |
ClinGen TOPMed |
|
|
CA292697207 rs954649959 |
39 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA400773984 rs954649959 |
39 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1451275791 CA400773985 |
39 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA400773986 rs1451275791 |
39 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA400774043 rs1402859766 |
47 | D>E | No |
ClinGen TOPMed |
|
|
rs1020476119 CA292697209 |
48 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1010565001 CA292697210 |
50 | K>* | No |
ClinGen TOPMed gnomAD |
|
|
CA292697235 rs968996691 |
52 | C>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA292697236 rs532612820 |
55 | Y>* | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs1421198808 CA400774104 |
55 | Y>H | No |
ClinGen gnomAD |
|
|
rs145659794 CA8686896 |
58 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs145659794 CA400774126 |
58 | P>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA400774155 rs551743110 |
62 | F>L | No |
ClinGen Ensembl |
|
|
CA292697242 rs201203253 |
64 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs772362348 CA8686898 COSM2793476 |
64 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA400774179 rs1340311056 |
66 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs999877578 CA292697243 |
67 | H>N | No |
ClinGen TOPMed |
|
|
rs1431930587 CA400774186 |
67 | H>Q | No |
ClinGen gnomAD |
|
|
CA400774190 rs1272190968 |
68 | L>P | No |
ClinGen gnomAD |
|
|
CA400774196 rs1360102990 |
69 | P>L | No |
ClinGen gnomAD |
|
|
rs1323079958 CA400774203 |
70 | R>S | No |
ClinGen gnomAD |
|
|
rs1033984387 CA292697244 |
71 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA400774217 rs1281164050 |
72 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400774219 rs1350051743 |
73 | P>A | No |
ClinGen gnomAD |
|
|
rs1284031165 CA400774223 COSM1135971 |
73 | P>L | kidney [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1350051743 CA400774220 |
73 | P>S | No |
ClinGen gnomAD |
|
|
CA400774225 rs1567907245 |
74 | Q>* | No |
ClinGen Ensembl |
|
|
rs760874803 CA8686901 |
74 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400774227 rs941296080 |
74 | Q>P | No |
ClinGen TOPMed gnomAD |
|
|
rs941296080 CA292697245 |
74 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA292697246 rs958446692 |
75 | K>N | No |
ClinGen TOPMed |
|
|
rs146549419 CA8686902 |
77 | K>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1216201404 CA400774260 |
79 | K>R | No |
ClinGen gnomAD |
|
|
rs1447319435 CA400774271 CA400774272 |
80 | Y>* | No |
ClinGen gnomAD |
|
|
rs1232834976 CA400774270 |
80 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA400774284 rs1187468594 |
82 | K>E | No |
ClinGen gnomAD |
|
|
rs1187468594 CA400774282 |
82 | K>Q | No |
ClinGen gnomAD |
|
|
CA400774292 rs1395173083 |
83 | Q>* | No |
ClinGen gnomAD |
|
|
rs1171411028 CA400774302 |
84 | S>I | No |
ClinGen TOPMed gnomAD |
|
|
rs750236898 CA8686903 |
85 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1401704247 CA400774311 |
86 | V>M | No |
ClinGen gnomAD |
|
|
rs1598333850 CA400774321 |
87 | V>A | No |
ClinGen Ensembl |
|
|
CA400774317 rs1320714766 |
87 | V>I | No |
ClinGen gnomAD |
|
| TCGA novel | 89 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8686904 rs555202197 |
90 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA292697249 rs984175020 |
90 | D>G | No |
ClinGen Ensembl |
|
|
CA292697248 rs565138902 |
90 | D>H | No |
ClinGen Ensembl |
|
|
CA8686905 rs181890967 |
91 | H>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA400774346 rs1244806327 |
92 | I>L | No |
ClinGen Ensembl |
|
|
CA400774361 rs1598333885 |
94 | T>P | No |
ClinGen Ensembl |
|
|
rs1232173222 CA400774386 |
97 | Q>H | No |
ClinGen gnomAD |
|
|
rs1050832950 CA292697250 |
99 | Y>* | No |
ClinGen TOPMed |
|
|
rs1288650442 CA400774409 |
100 | C>* | No |
ClinGen gnomAD |
|
|
CA400774424 rs1489733012 |
102 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1260173974 CA400774419 |
102 | A>T | No |
ClinGen TOPMed |
|
|
CA400774423 rs1489733012 |
102 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA400774428 rs1273046095 |
103 | W>* | No |
ClinGen gnomAD |
|
|
rs1598333914 CA400774436 |
104 | E>* | No |
ClinGen Ensembl |
|
|
CA292697251 rs892209606 |
106 | K>E | No |
ClinGen TOPMed |
|
|
CA400774452 rs1205608606 |
106 | K>R | No |
ClinGen gnomAD |
|
|
CA292697252 rs1010401198 |
107 | H>Y | No |
ClinGen TOPMed |
|
|
rs1286090759 CA400774464 |
108 | W>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1463334079 CA400774474 |
109 | R>K | No |
ClinGen gnomAD |
|
|
rs766242804 CA8686922 |
111 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762682105 CA8686921 |
111 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs774272052 CA8686923 |
112 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400774515 rs1293709437 |
113 | W>* | No |
ClinGen gnomAD |
|
|
CA400774518 rs1323216308 |
113 | W>C | No |
ClinGen gnomAD |
|
|
rs759533511 CA8686924 |
114 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs935939750 CA292697296 |
114 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA400774525 rs1598334291 |
115 | F>V | No |
ClinGen Ensembl |
|
|
CA400774532 rs1598334298 |
116 | I>V | No |
ClinGen Ensembl |
|
|
CA8686927 rs755759396 |
117 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs894166861 CA292697297 |
118 | L>F | No |
ClinGen Ensembl |
|
|
rs763684116 CA8686928 |
120 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs372731739 CA8686929 |
120 | S>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8686932 rs746364850 |
123 | E>* | No |
ClinGen ExAC gnomAD |
|
|
CA8686933 rs759024303 |
126 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs1028238174 CA292697299 |
126 | Q>R | No |
ClinGen TOPMed |
|
|
CA8686934 rs780153529 |
128 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA8686935 rs747093678 |
128 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA8686936 rs768877828 |
129 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs776726903 CA400774612 |
129 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs776726903 CA8686937 |
129 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1165498733 CA400774615 |
130 | L>F | No |
ClinGen TOPMed |
|
|
CA400774622 rs1381145488 |
131 | Y>F | No |
ClinGen gnomAD |
|
|
CA400774660 rs1326631728 |
136 | S>N | No |
ClinGen gnomAD |
|
|
rs376243760 CA8686941 |
137 | S>R | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1028549273 CA292697301 |
138 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs868758149 CA292697302 |
139 | P>A | No |
ClinGen gnomAD |
|
|
CA8686942 rs560519932 |
139 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs868758149 CA292697303 |
139 | P>S | No |
ClinGen gnomAD |
|
|
rs760125686 CA8686944 |
141 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA8686945 rs763735613 |
141 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1262814333 CA400774690 |
141 | K>T | No |
ClinGen gnomAD |
|
|
rs753429551 CA8686946 |
142 | F>C | No |
ClinGen ExAC |
|
|
rs763653945 CA292697305 |
142 | F>L | No |
ClinGen Ensembl |
|
|
CA8686981 rs779956263 |
144 | V>G | No |
ClinGen ExAC gnomAD |
|
|
CA8686980 rs771760090 |
144 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA400774914 rs746897514 |
146 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs768677332 CA8686984 |
147 | C>G | No |
ClinGen ExAC gnomAD |
|
|
CA8686983 rs768677332 |
147 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs769510201 CA8686987 |
148 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8686986 rs769510201 |
148 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400774931 rs1408484761 |
149 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1230302825 CA400774945 |
151 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8686991 rs57928695 |
152 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA400774948 rs1333258221 |
152 | P>S | No |
ClinGen gnomAD |
|
|
rs752725736 CA8686993 |
153 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs140000868 CA292697595 |
154 | R>M | No |
ClinGen ESP |
|
|
rs1598336260 CA400774964 |
155 | S>A | No |
ClinGen Ensembl |
|
|
rs756200782 CA8686994 |
155 | S>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA8686996 rs754046740 |
156 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs201450863 CA8686999 |
159 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs768426490 CA400774996 |
160 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8687000 rs768426490 |
160 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA400775002 rs1426418826 |
161 | E>G | No |
ClinGen TOPMed |
|
|
CA292697597 rs944359779 |
162 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA8687002 rs747591049 |
162 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1352020133 CA400775029 |
165 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1352020133 CA400775028 |
165 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1200441763 CA400775055 |
169 | H>R | No |
ClinGen gnomAD |
|
|
CA8687004 rs200962057 |
169 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400775066 rs1450596616 |
171 | W>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA8687008 rs574216649 |
172 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs574216649 CA292697599 |
172 | G>E | No |
ClinGen ExAC gnomAD |
|
|
CA292697600 rs760760620 |
174 | H>Q | No |
ClinGen TOPMed |
|
|
rs1378750483 CA400775087 |
174 | H>R | No |
ClinGen gnomAD |
|
|
CA292697601 rs370364055 |
175 | S>G | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA400775100 rs1462880494 |
176 | N>S | No |
ClinGen gnomAD |
|
|
CA400775113 rs1299927823 |
178 | S>G | No |
ClinGen gnomAD |
|
|
CA8687010 rs368635684 |
179 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8687011 rs775980827 |
180 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761228169 CA8687012 |
181 | K>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA292697602 rs1003725055 |
182 | E>* | No |
ClinGen TOPMed |
|
|
rs1056158164 CA292697603 |
182 | E>G | No |
ClinGen Ensembl |
|
|
CA8687013 rs572279125 |
183 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA400775147 COSM1563720 rs1417608936 |
183 | R>W | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA8687015 rs757380106 |
185 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs1027739290 CA292697604 |
186 | N>K | No |
ClinGen TOPMed gnomAD |
|
|
rs751251927 CA292697605 |
188 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA8687017 rs751251927 |
188 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA400775177 rs1567908955 |
188 | Q>R | No |
ClinGen Ensembl |
|
|
rs1598336422 CA400775182 |
189 | T>P | No |
ClinGen Ensembl |
|
|
rs371681055 CA8687018 |
190 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1254833362 CA400775196 |
191 | G>D | No |
ClinGen TOPMed |
|
| TCGA novel | 192 | Q>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8687019 rs539594178 |
194 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1332647455 CA400775227 |
196 | L>M | No |
ClinGen gnomAD |
|
|
rs1332647455 CA400775226 |
196 | L>V | No |
ClinGen gnomAD |
|
|
rs1007801819 CA292697607 |
199 | Q>K | No |
ClinGen Ensembl |
|
| TCGA novel | 199 | Q>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1189425069 CA400775259 |
201 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1436108665 CA400775271 |
203 | Q>* | No |
ClinGen gnomAD |
|
|
rs1272236644 CA400775284 |
204 | D>E | No |
ClinGen TOPMed |
|
|
rs1322796589 CA400775280 |
204 | D>G | No |
ClinGen gnomAD |
|
|
rs1363194769 CA400775287 |
205 | H>Y | No |
ClinGen gnomAD |
|
|
CA292697608 rs201950282 |
209 | D>H | No |
ClinGen TOPMed gnomAD |
|
|
CA400775631 COSM3421743 rs1456917549 |
210 | S>Y | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA400775636 rs1163992457 |
211 | L>S | No |
ClinGen gnomAD |
|
|
rs777790196 CA8687045 |
212 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8687046 rs748690148 |
214 | L>R | No |
ClinGen ExAC gnomAD |
|
|
rs1403672629 CA400775655 |
214 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8687047 rs756797485 |
215 | S>P | No |
ClinGen ExAC gnomAD |
|
|
rs888518494 CA292697816 |
216 | Q>* | No |
ClinGen gnomAD |
|
| TCGA novel | 217 | K>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400775681 rs1237956925 |
218 | P>Q | No |
ClinGen gnomAD |
|
|
CA400775682 rs1237956925 |
218 | P>R | No |
ClinGen gnomAD |
|
|
CA292697817 rs202131111 |
218 | P>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes NCI-TCGA TOPMed gnomAD |
|
rs1357902675 CA400775693 |
220 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs771751554 CA8687050 |
221 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400775707 rs1037498164 |
222 | R>K | No |
ClinGen gnomAD |
|
|
CA292697818 rs1037498164 |
222 | R>M | No |
ClinGen gnomAD |
|
|
rs747395163 CA8687052 |
225 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400775729 rs1250080135 |
225 | I>T | No |
ClinGen gnomAD |
|
|
rs1472460407 CA400775748 |
228 | A>T | No |
ClinGen gnomAD |
|
|
rs769091687 CA8687053 |
228 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA8687054 rs777196412 |
231 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA292697819 rs756919587 |
233 | E>D | No |
ClinGen Ensembl |
|
| TCGA novel | 234 | T>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400775791 rs1371172838 |
235 | E>K | No |
ClinGen gnomAD |
|
|
CA8687059 rs150310077 |
236 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs370226193 CA8687058 |
236 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs137972332 CA8687061 |
237 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
No associated diseases with Q86WR6
1 regional properties for Q86WR6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Chromodomain-helicase-DNA-binding protein 1-like, C-terminal domain | 31 - 134 | IPR025260 |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
3 homologous proteins in AiPD
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEASDGQGGE | GDKPLEQVTN | VSCLETSSSA | SPARDSLMRH | AKGLDQDTFK | TCKEYLRPLK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KFLRKLHLPR | DLPQKKKLKY | MKQSLVVLGD | HINTFLQHYC | QAWEIKHWRK | MLWRFISLFS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ELEAKQLRRL | YKYTKSSQPA | KFLVTFCASD | APERSLLADR | EDSLPKLCHA | WGLHSNISGM |
| 190 | 200 | 210 | 220 | 230 | |
| KERLSNMQTP | GQGSPLPGQP | RSQDHVKKDS | LRELSQKPKL | KRKRIKEAPE | TPETEP |