Q86WJ1
Gene name |
CHD1L (ALC1) |
Protein name |
Chromodomain-helicase-DNA-binding protein 1-like |
Names |
EC 3.6.4.- , Amplified in liver cancer protein 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9557 |
EC number |
3.6.4.-: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1-LIKE (PTHR47157) |
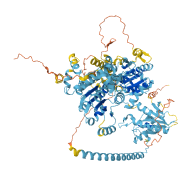
Descriptions
Oncogenic chromatin remodeler ALC1 (ALC1, also known as CHD1L) is an ATP-dependent chromatin remodeler that relaxes the chromatin and plays an important role in the poly(ADP-ribose) polymerase 1-mediated DNA repair pathway. The macro domain of ALC1 binds to lobe 2 of the ATPase motor, sequestering two elements for nucleosome recognition. Autoinhibition by macro domain interactions stabilizes the ATPase lobe, impacting DNA damage repair. The cancer-related mutations of several basic residues on the macro domain disrupt the autoinhibition of ALC1, compromising its function in cells. Upon DNA damage, the interaction between the macro domain and PAR chains in PARP1 can also release the autoinhibition of ALC1 and target the remodeler to the sites of DNA damage. Furthermore, the histone H4 tails of nucleosome and other nucleosomal epitopes often provide the combinational cues to target and regulate chromatin remodelers, leading to the activation of ALC1 in cells.
Autoinhibitory domains (AIDs)
Target domain |
348-513 (Lobe 2 of ATPase motor domain) |
Relief mechanism |
Ligand binding, Partner binding, Others |
Assay |
Structural analysis, Mutagenesis experiment, Peptide inhibitor test, Deletion assay |
Accessory elements
No accessory elements
References
Autoinhibited structure
Activated structure
1100 variants for Q86WJ1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000416565 CA1063384 rs201333837 RCV000994092 |
236 | R>H | Congenital anomaly of kidney and urinary tract [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001374538 rs782747449 |
419 | L>P | Hereditary breast ovarian cancer syndrome [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs376783101 RCV003418384 RCV002246682 RCV002137548 |
517 | I>M | CHD1L-related condition [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs1064797118 RCV000488392 RCV003278839 CA16621573 |
629 | T>N | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
CA1063983 rs149913688 RCV000484211 RCV002526964 |
727 | P>T | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs369564534 RCV000416573 CA1063991 |
736 | I>V | Congenital anomaly of kidney and urinary tract [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000416594 CA1064038 rs782687350 |
760 | A>T | Congenital anomaly of kidney and urinary tract [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000913227 rs147494800 |
1 | M>L | No |
ClinVar dbSNP |
|
| rs782190965 | 2 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs377310969 | 3 | R>C | No |
ESP TOPMed gnomAD |
|
| rs1255079978 | 3 | R>H | No | TOPMed | |
| rs782305182 | 4 | A>P | No |
1000Genomes ExAC gnomAD |
|
| rs1665082125 | 4 | A>V | No | TOPMed | |
| rs1222127022 | 5 | G>A | No |
TOPMed gnomAD |
|
| rs782387057 | 5 | G>R | No |
ExAC gnomAD |
|
| rs1665087556 | 6 | A>G | No | Ensembl | |
| rs782153707 | 6 | A>T | No |
ExAC gnomAD |
|
| rs1447633273 | 7 | T>A | No |
TOPMed gnomAD |
|
| rs1571513645 | 7 | T>I | No | Ensembl | |
| rs1447633273 | 7 | T>S | No |
TOPMed gnomAD |
|
| rs373271315 | 8 | S>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373271315 | 8 | S>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782101307 | 8 | S>R | No | ExAC | |
| rs782736815 | 9 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs782736815 | 9 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs782506295 | 9 | R>H | No |
ExAC TOPMed |
|
| rs782506295 | 9 | R>P | No |
ExAC TOPMed |
|
| rs781885928 | 10 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs782465698 | 10 | G>V | No |
ExAC gnomAD |
|
| rs781885928 | 10 | G>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 11 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782225008 | 11 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs782341713 | 11 | G>D | No |
ExAC gnomAD |
|
| rs782225008 | 11 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs782341713 | 11 | G>V | No |
ExAC gnomAD |
|
| rs782646191 | 12 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs782646191 | 12 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs587765929 | 12 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782646191 | 12 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs782416368 | 12 | Q>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
RCV001639477 rs1890042 |
13 | A>= | No |
ClinVar dbSNP |
|
| rs782133558 | 13 | A>D | No |
ExAC gnomAD |
|
| rs782370058 | 13 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs782370058 | 13 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1280350026 | 14 | P>R | No |
TOPMed gnomAD |
|
| rs1665113158 | 15 | G>S | No | TOPMed | |
| rs782152009 | 16 | F>Y | No |
ExAC TOPMed gnomAD |
|
| rs1553931845 | 17 | L>I | No | gnomAD | |
| rs1553931853 | 19 | R>Q | No | gnomAD | |
| rs1665119164 | 20 | L>F | No | Ensembl | |
| rs1665120827 | 21 | H>P | No | TOPMed | |
| rs1439164074 | 21 | H>Q | No | TOPMed | |
| rs1665120827 | 21 | H>R | No | TOPMed | |
| rs1351011184 | 22 | T>A | No |
TOPMed gnomAD |
|
| rs1665123252 | 22 | T>I | No | TOPMed | |
| rs1665124972 | 23 | E>D | No | TOPMed | |
| rs1553931874 | 24 | G>D | No | gnomAD | |
| rs1391823347 | 25 | R>* | No | TOPMed | |
| rs11588753 | 25 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs11588753 RCV001666999 VAR_042954 |
25 | R>P | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs11588753 | 25 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1665130885 | 27 | E>K | No | gnomAD | |
| rs1665131566 | 27 | E>V | No | gnomAD | |
| rs200287122 | 28 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1213085797 | 29 | A>E | No |
TOPMed gnomAD |
|
| rs1213085797 | 29 | A>V | No |
TOPMed gnomAD |
|
| rs782676436 | 30 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs972230040 | 30 | R>Q | No |
TOPMed gnomAD |
|
| rs782676436 | 30 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1293833748 | 32 | Q>* | No | gnomAD | |
| rs1221302265 | 32 | Q>L | No | Ensembl | |
| rs1665141310 | 33 | E>K | No |
TOPMed gnomAD |
|
| rs1368853275 | 34 | Q>K | No |
TOPMed gnomAD |
|
| rs587687001 | 35 | D>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782618260 | 37 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1665145713 | 37 | R>P | No | TOPMed | |
| rs781889081 | 38 | Q>K | No | Ensembl | |
| rs1332924827 | 39 | W>C | No |
TOPMed gnomAD |
|
| rs930822717 | 40 | G>A | No |
TOPMed gnomAD |
|
| TCGA novel | 42 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782246341 | 44 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs782246341 | 44 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs945236631 | 45 | H>Y | No | gnomAD | |
| rs1455863597 | 47 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1553937191 | 47 | R>H | No |
TOPMed gnomAD |
|
| rs1668775921 | 48 | S>A | No | Ensembl | |
| rs782200038 | 49 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs1553937204 | 49 | Y>H | No |
TOPMed gnomAD |
|
| rs1042273506 | 50 | Q>* | No | gnomAD | |
| rs1471983424 | 50 | Q>H | No |
TOPMed gnomAD |
|
| rs782311405 | 52 | E>D | No |
ExAC gnomAD |
|
| TCGA novel | 52 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781965410 | 53 | G>E | No |
ExAC gnomAD |
|
| rs587755825 | 54 | V>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs587755825 | 54 | V>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200995927 | 55 | N>D | No | Ensembl | |
| rs587645225 | 56 | W>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 56 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782383196 | 56 | W>R | No |
ExAC gnomAD |
|
| rs1553937244 | 57 | L>F | No | gnomAD | |
| rs782803495 | 58 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs782803495 | 58 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1553937281 | 59 | Q>H | No | gnomAD | |
| rs1553937264 | 59 | Q>P | No | gnomAD | |
| rs1553937264 | 59 | Q>R | No | gnomAD | |
|
COSM423714 rs782045573 COSM5207606 |
60 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs782737857 | 60 | R>H | No |
ExAC TOPMed gnomAD |
|
|
COSM895280 rs587758592 |
62 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 64 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3473183 | 65 | N>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1668797085 | 65 | N>S | No | Ensembl | |
| rs1668797839 | 66 | G>S | No | gnomAD | |
| rs782470880 | 67 | C>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 69 | L>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 69 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM895281 | 70 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1553937308 | 72 | E>G | No |
TOPMed gnomAD |
|
| rs782806343 | 72 | E>K | No |
ExAC gnomAD |
|
| rs1234172839 | 74 | G>D | No |
TOPMed gnomAD |
|
| rs1234172839 | 74 | G>V | No |
TOPMed gnomAD |
|
| rs1553937318 | 76 | G>W | No | gnomAD | |
| rs1668802904 | 77 | K>* | No | gnomAD | |
| rs1553937327 | 78 | T>A | No | gnomAD | |
| rs1553937335 | 78 | T>I | No | Ensembl | |
| rs1553937327 | 78 | T>P | No | gnomAD | |
| rs1553937335 | 78 | T>S | No | Ensembl | |
| rs2102364378 | 79 | C>G | No | Ensembl | |
| rs2102364410 | 80 | Q>E | No | Ensembl | |
| rs781889467 | 80 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1553938883 | 81 | T>A | No | gnomAD | |
|
COSM5176803 COSM1333651 |
81 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1382345863 | 82 | I>T | No |
TOPMed gnomAD |
|
| rs1571695588 | 82 | I>V | No |
TOPMed gnomAD |
|
| rs1669578407 | 83 | A>P | No | Ensembl | |
| rs36068896 | 84 | L>F | No |
TOPMed gnomAD |
|
| rs36068896 | 84 | L>V | No |
TOPMed gnomAD |
|
| rs1559749560 | 85 | F>L | No | Ensembl | |
| rs1425106117 | 86 | I>M | No | TOPMed | |
| rs782051281 | 86 | I>V | No |
ExAC gnomAD |
|
| rs1669585037 | 87 | Y>S | No | Ensembl | |
| rs369003776 | 88 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs781953321 | 88 | L>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 89 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1011097726 | 90 | G>E | No | Ensembl | |
| rs1669588360 | 91 | R>S | No | Ensembl | |
| rs371676695 | 92 | L>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM3473184 | 92 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782145750 | 96 | G>V | No |
ExAC gnomAD |
|
| rs781905122 | 96 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs782712681 | 97 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs782712681 | 97 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs782648240 | 98 | F>L | No |
ExAC gnomAD |
|
| rs781862870 | 101 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs370027194 | 103 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1669599810 | 110 | W>R | No | gnomAD | |
| rs1343665947 | 111 | K>R | No |
TOPMed gnomAD |
|
| rs1669922224 | 117 | F>C | No |
TOPMed gnomAD |
|
| rs1553939837 | 118 | A>P | No | gnomAD | |
| rs1669924526 | 118 | A>V | No | TOPMed | |
| rs782474557 | 119 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1669926283 | 119 | P>Q | No | TOPMed | |
| rs1669926283 | 119 | P>R | No | TOPMed | |
| rs782474557 | 119 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1669928060 | 120 | G>D | No | TOPMed | |
| rs1553939855 | 120 | G>S | No | gnomAD | |
| rs1226320177 | 121 | L>V | No | TOPMed | |
| rs1553939864 | 123 | C>Y | No | gnomAD | |
| rs782243350 | 124 | V>E | No |
ExAC gnomAD |
|
| rs1669931031 | 124 | V>I | No | gnomAD | |
|
rs1669933364 COSM3862599 |
125 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1669934168 | 125 | T>I | No |
TOPMed gnomAD |
|
| rs201227166 | 126 | Y>H | No |
TOPMed gnomAD |
|
| rs782540038 | 127 | A>V | No | ExAC | |
| rs1669939393 | 129 | D>G | No | TOPMed | |
| rs1344772841 | 129 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs201990873 | 130 | K>R | No | gnomAD | |
| rs146325614 | 132 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs782306437 | 133 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs781957384 | 133 | R>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 133 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs192336187 | 134 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1669947100 | 135 | C>Y | No | Ensembl | |
| rs1553939941 | 136 | L>F | No | gnomAD | |
| rs1559753887 | 136 | L>P | No | TOPMed | |
| rs1553939952 | 138 | Q>P | No | gnomAD | |
| rs910575786 | 139 | D>G | No | Ensembl | |
| rs1553939959 | 139 | D>N | No | gnomAD | |
| rs201680696 | 145 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782037968 | 145 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1177553313 | 147 | H>R | No | TOPMed | |
| rs372855761 | 148 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs372855761 | 148 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1669961788 | 149 | L>Q | No | TOPMed | |
| rs1184674375 | 151 | T>I | No | Ensembl | |
| rs1553940014 | 152 | T>A | No | gnomAD | |
|
rs1669968602 RCV001591755 |
153 | Y>* | No |
ClinVar TOPMed dbSNP |
|
| rs782730027 | 153 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs782730027 | 153 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs782146681 | 155 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs782146681 | 155 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1285620240 | 156 | C>F | No |
TOPMed gnomAD |
|
| rs1670214624 | 156 | C>W | No | TOPMed | |
| rs587719271 | 157 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1670218552 | 159 | D>G | No | Ensembl | |
| rs1670217835 | 159 | D>N | No | Ensembl | |
| rs1375587977 | 160 | A>S | No |
TOPMed gnomAD |
|
| rs1375587977 | 160 | A>T | No |
TOPMed gnomAD |
|
| rs1454200114 | 160 | A>V | No |
TOPMed gnomAD |
|
| COSM3473185 | 161 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs909378755 | 162 | F>C | No | Ensembl | |
| rs1670223534 | 163 | L>P | No | TOPMed | |
|
COSM5480871 COSM5480870 rs1379475915 |
165 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs781849750 | 166 | F>C | No |
ExAC gnomAD |
|
| rs1553942722 | 167 | P>L | No |
TOPMed gnomAD |
|
| rs782465288 | 168 | W>* | No |
ExAC gnomAD |
|
| rs1553942729 | 168 | W>* | No | gnomAD | |
| rs782465288 | 168 | W>C | No |
ExAC gnomAD |
|
| rs1553942742 | 169 | S>I | No | gnomAD | |
| rs371872866 | 170 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4402558 COSM4402559 rs1671331917 |
171 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| TCGA novel | 171 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1231736748 | 172 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1470056996 | 172 | V>L | No | TOPMed | |
| rs782562082 | 173 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1553942782 | 173 | V>L | No | gnomAD | |
| rs1553942793 | 174 | D>E | No | gnomAD | |
| TCGA novel | 175 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782199607 | 178 | R>K | No |
ExAC TOPMed gnomAD |
|
| COSM1127405 | 179 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782439013 | 179 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1671342555 | 179 | L>W | No | TOPMed | |
| rs782610778 | 181 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1553942814 | 183 | S>T | No | Ensembl | |
| rs1671344902 | 184 | S>F | No | gnomAD | |
| rs1286047552 | 185 | L>R | No |
TOPMed gnomAD |
|
| rs1224176633 | 186 | L>M | No |
TOPMed gnomAD |
|
| rs1372508809 | 187 | H>D | No |
TOPMed gnomAD |
|
| rs202041182 | 187 | H>R | No |
1000Genomes ExAC gnomAD |
|
| rs782381218 | 188 | K>E | No |
ExAC gnomAD |
|
| rs1553942835 | 188 | K>R | No | gnomAD | |
| rs1671353639 | 190 | L>S | No | Ensembl | |
| rs1671354451 | 191 | S>* | No |
TOPMed gnomAD |
|
|
COSM260088 COSM5098755 |
193 | F>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781957034 | 194 | S>* | No | ExAC | |
| rs782366790 | 194 | S>A | No |
ExAC gnomAD |
|
| rs1673111080 | 195 | V>E | No | TOPMed | |
| rs782265673 | 196 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs782265673 | 196 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1553946467 | 198 | S>G | No | gnomAD | |
| rs1673117633 | 199 | L>H | No | TOPMed | |
| rs1673116129 | 199 | L>V | No | TOPMed | |
| rs1553946490 | 201 | L>* | No | gnomAD | |
| rs782724329 | 201 | L>F | No |
ExAC gnomAD |
|
| rs587674066 | 202 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1458109974 | 203 | G>E | No | TOPMed | |
| rs148845749 | 203 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1237494067 | 204 | T>S | No |
TOPMed gnomAD |
|
| rs1196894937 | 206 | I>L | No |
TOPMed gnomAD |
|
| rs1673128078 | 207 | Q>* | No |
TOPMed gnomAD |
|
| rs1553946535 | 207 | Q>H | No | gnomAD | |
| rs1673128870 | 207 | Q>R | No | gnomAD | |
| rs143518204 | 208 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1553946543 | 210 | L>F | No | gnomAD | |
| rs1673138959 | 212 | E>K | No | Ensembl | |
| rs782808030 | 213 | L>P | No |
ExAC gnomAD |
|
| rs1553946566 | 214 | Y>* | No | gnomAD | |
| rs1281579709 | 214 | Y>C | No |
TOPMed gnomAD |
|
| rs1553946571 | 215 | S>F | No | Ensembl | |
| rs1571838548 | 219 | F>V | No | Ensembl | |
| rs1673148166 | 221 | E>G | No | TOPMed | |
| rs1673149031 | 222 | P>A | No | Ensembl | |
| rs587632403 | 222 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs955249999 | 224 | L>P | No |
TOPMed gnomAD |
|
| rs1553946614 | 226 | S>F | No | gnomAD | |
| rs781826569 | 226 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs782520618 | 227 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 229 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782636756 | 229 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs973398344 | 230 | V>G | No | Ensembl | |
| rs1673159802 | 230 | V>M | No |
TOPMed gnomAD |
|
| rs1553946638 | 231 | G>E | No |
TOPMed gnomAD |
|
| rs1379671655 | 233 | F>V | No |
TOPMed gnomAD |
|
| rs1673165244 | 235 | Q>* | No | TOPMed | |
| rs782408250 | 235 | Q>H | No |
ExAC gnomAD |
|
| rs587768473 | 236 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs782354202 | 239 | D>Y | No |
ExAC TOPMed gnomAD |
|
|
COSM5514784 COSM5514783 |
243 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781953345 | 243 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs782753018 | 244 | S>C | No | Ensembl | |
| rs2102547604 | 244 | S>P | No | Ensembl | |
| rs139274984 | 245 | E>D | No |
ESP TOPMed gnomAD |
|
| rs1673177775 | 245 | E>Q | No | Ensembl | |
| rs199929968 | 247 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1553947848 | 248 | S>N | No | gnomAD | |
| rs1673738391 | 248 | S>R | No | TOPMed | |
| rs1553947848 | 248 | S>T | No | gnomAD | |
| rs781925575 | 249 | E>K | No |
ExAC gnomAD |
|
| rs1417974414 | 250 | L>V | No | TOPMed | |
| rs1553947868 | 251 | H>R | No | gnomAD | |
| rs1571856991 | 252 | K>N | No | Ensembl | |
| rs782107935 | 254 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs587612852 | 254 | L>S | No | 1000Genomes | |
| rs587612852 | 254 | L>W | No | 1000Genomes | |
| rs782344210 | 255 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs1673750947 | 256 | P>L | No | Ensembl | |
| rs1553947918 | 257 | F>S | No |
TOPMed gnomAD |
|
| rs1256469276 | 258 | L>Q | No |
TOPMed gnomAD |
|
| rs782819498 | 259 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs782819498 | 259 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1673755990 | 260 | R>M | No | gnomAD | |
| TCGA novel | 260 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs144288940 | 261 | R>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs144288940 | 261 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs202226947 | 261 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202226947 | 261 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1553947957 | 262 | V>M | No |
TOPMed gnomAD |
|
| rs1553947977 | 263 | K>N | No | gnomAD | |
| rs1315139999 | 264 | A>P | No |
TOPMed gnomAD |
|
| rs1229050451 | 265 | E>D | No |
TOPMed gnomAD |
|
| rs1553947987 | 269 | E>D | No |
TOPMed gnomAD |
|
| rs1355232999 | 272 | K>E | No | TOPMed | |
| rs1553948000 | 272 | K>N | No |
TOPMed gnomAD |
|
| rs1673772258 | 272 | K>R | No | TOPMed | |
| rs782770115 | 273 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs781848220 | 274 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs112618269 | 274 | T>I | No | TOPMed | |
| rs781848220 | 274 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1028414868 | 276 | V>I | No |
TOPMed gnomAD |
|
| rs782537728 | 277 | V>M | No |
ExAC gnomAD |
|
|
RCV000385994 CA10602731 rs886041164 |
278 | I>M | No |
ClinGen ClinVar TOPMed dbSNP |
|
| rs782494149 | 279 | Y>C | No |
ExAC gnomAD |
|
| rs782669205 | 279 | Y>D | No |
ExAC gnomAD |
|
| rs782669205 | 279 | Y>H | No |
ExAC gnomAD |
|
| rs1673785139 | 281 | G>D | No | Ensembl | |
| TCGA novel | 282 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs587737412 | 283 | S>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs587737412 | 283 | S>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs587737412 | 283 | S>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1571858647 | 284 | A>T | No | Ensembl | |
| rs2102572127 | 285 | L>F | No | Ensembl | |
| rs1416708695 | 285 | L>S | No |
TOPMed gnomAD |
|
| rs2102572170 | 286 | Q>K | No | Ensembl | |
| rs1673795170 | 288 | K>* | No | TOPMed | |
| rs1185594393 | 288 | K>I | No | TOPMed | |
| rs1185594393 | 288 | K>R | No | TOPMed | |
| rs200436767 | 289 | Y>H | No |
1000Genomes gnomAD |
|
| rs1553948134 | 290 | Y>* | No | gnomAD | |
| rs1553948121 | 290 | Y>H | No | gnomAD | |
| rs1262381281 | 292 | A>T | No |
TOPMed gnomAD |
|
| rs1553948161 | 294 | L>M | No | gnomAD | |
| rs1673806412 | 295 | M>I | No | Ensembl | |
| rs781926083 | 295 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1673807346 | 296 | K>E | No | Ensembl | |
| rs782098154 | 297 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs782728909 | 299 | D>G | No | TOPMed | |
| rs782027400 | 300 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1210725411 | 300 | A>V | No | Ensembl | |
| COSM98080 | 301 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs369279428 | 301 | F>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs782317795 | 302 | E>G | No |
ExAC gnomAD |
|
| rs587697021 | 303 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1674387739 | 304 | E>D | No | Ensembl | |
| rs1553949286 | 304 | E>K | No | gnomAD | |
| rs782085796 | 305 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs782085796 | 305 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1553949300 | 307 | K>Q | No |
TOPMed gnomAD |
|
| rs1674395055 | 308 | K>E | No | Ensembl | |
| rs1571876795 | 309 | V>A | No | Ensembl | |
|
COSM674768 rs1674398726 |
312 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs782452148 | 312 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs781808589 | 312 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs781808589 | 312 | Q>R | No |
ExAC TOPMed gnomAD |
|
|
COSM1333652 COSM5165455 |
313 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782614177 | 314 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1674404784 | 317 | Q>* | No | Ensembl | |
|
COSM4671952 rs587754399 COSM4671951 |
319 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs202245685 | 319 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs202245685 | 319 | R>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 320 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1674408003 | 322 | V>L | No | TOPMed | |
| rs782634856 | 323 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs782634856 | 323 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs2102597202 | 323 | D>Y | No | Ensembl | |
| rs1553949378 | 324 | H>Y | No | gnomAD | |
| rs373036183 | 328 | F>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs373036183 | 328 | F>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1418948373 | 329 | D>H | No |
TOPMed gnomAD |
|
| rs1418948373 | 329 | D>N | No |
TOPMed gnomAD |
|
| rs1553949419 | 330 | G>C | No | gnomAD | |
| rs782329452 | 330 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs782349484 | 331 | V>G | No |
ExAC gnomAD |
|
| rs782235742 | 331 | V>M | No |
ExAC gnomAD |
|
| rs1249494722 | 333 | P>A | No |
TOPMed gnomAD |
|
| rs782811744 | 333 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs782811744 | 333 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1249494722 | 333 | P>T | No |
TOPMed gnomAD |
|
| TCGA novel | 335 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1674977041 | 336 | F>V | No | TOPMed | |
| rs782009110 | 338 | V>I | No |
ExAC gnomAD |
|
| rs1553950500 | 339 | G>E | No | gnomAD | |
| rs369233757 | 340 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs140168816 | 340 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM3473186 | 340 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781839697 | 341 | H>P | No |
ExAC TOPMed gnomAD |
|
| rs781839697 | 341 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1553950514 | 341 | H>Y | No | gnomAD | |
| COSM895283 | 343 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2102620427 | 343 | T>I | No | Ensembl | |
| rs1553950539 | 343 | T>P | No | gnomAD | |
| rs373166478 | 344 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs782557151 | 346 | S>N | No |
TOPMed gnomAD |
|
| rs782557151 | 346 | S>T | No |
TOPMed gnomAD |
|
| rs781903935 | 348 | K>R | No |
ExAC gnomAD |
|
| rs782546588 | 349 | L>F | No |
ExAC gnomAD |
|
|
RCV001723225 VAR_042955 rs17356233 |
350 | H>Q | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD ClinVar |
|
| rs1674998098 | 351 | L>V | No | Ensembl | |
| rs1229550550 | 353 | D>G | No |
TOPMed gnomAD |
|
| rs938650845 | 354 | K>Q | No | Ensembl | |
| rs149891977 | 357 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1301744095 | 357 | A>S | No |
TOPMed gnomAD |
|
| rs149891977 | 357 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs782259785 | 358 | F>C | No |
ExAC gnomAD |
|
| rs377698659 | 361 | S>P | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 361 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1362018070 | 362 | G>V | No | TOPMed | |
| rs1553952466 | 364 | H>R | No |
TOPMed gnomAD |
|
| TCGA novel | 364 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1024396953 | 365 | R>Q | No |
TOPMed gnomAD |
|
| rs368115096 | 365 | R>W | No |
ESP TOPMed gnomAD |
|
|
rs1675947168 RCV001092897 |
366 | V>A | No |
ClinVar Ensembl dbSNP |
|
| rs1238308009 | 366 | V>F | No |
TOPMed gnomAD |
|
| rs1361822490 | 368 | L>V | No | TOPMed | |
| rs1316209486 | 369 | F>C | No |
TOPMed gnomAD |
|
| rs1316209486 | 369 | F>S | No |
TOPMed gnomAD |
|
| rs1244459638 | 371 | Q>* | No | TOPMed | |
| rs782161166 | 373 | T>I | No |
ExAC gnomAD |
|
| rs782161166 | 373 | T>N | No |
ExAC gnomAD |
|
| rs1675955768 | 374 | Q>H | No | TOPMed | |
| rs1339242686 | 377 | D>V | No | Ensembl | |
| rs1320779160 | 378 | I>M | No |
TOPMed gnomAD |
|
| rs1675966121 | 380 | Q>H | No | Ensembl | |
| rs1385125557 | 380 | Q>R | No | TOPMed | |
| rs1553952556 | 381 | D>G | No | gnomAD | |
| rs144757186 | 381 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1417513790 | 382 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs201579599 | 383 | M>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1559811355 | 383 | M>L | No | Ensembl | |
| rs1386652451 | 383 | M>T | No |
TOPMed gnomAD |
|
| rs1553952585 | 385 | Y>F | No | gnomAD | |
| rs782525631 | 386 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs782687691 | 386 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1676496211 | 387 | G>A | No | TOPMed | |
| rs1195313919 | 387 | G>S | No | TOPMed | |
| rs199831401 | 390 | Y>C | No |
1000Genomes ExAC gnomAD |
|
| rs782609645 | 391 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs367735711 | 391 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs138710865 | 392 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs782668695 | 392 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs782668695 | 392 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs782668695 | 392 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs782323523 | 395 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1457716456 | 397 | V>L | No | TOPMed | |
| rs781913199 | 399 | G>R | No | ExAC | |
| rs782231747 | 400 | E>G | No |
ExAC TOPMed |
|
| rs1443709118 | 402 | R>K | No | TOPMed | |
| TCGA novel | 402 | R>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1278675848 | 403 | H>D | No | TOPMed | |
| rs1676520424 | 405 | A>S | No | Ensembl | |
| TCGA novel | 405 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1553953881 | 406 | I>V | No | gnomAD | |
| rs1676522308 | 407 | K>N | No | TOPMed | |
| rs1218327685 | 409 | F>C | No | TOPMed | |
| rs1676524928 | 409 | F>L | No | Ensembl | |
| rs1676525877 | 410 | G>R | No | Ensembl | |
| rs1345233141 | 411 | Q>L | No |
TOPMed gnomAD |
|
| rs1345233141 | 411 | Q>R | No |
TOPMed gnomAD |
|
| rs1676528446 | 412 | Q>* | No | TOPMed | |
| rs1553953908 | 412 | Q>R | No | gnomAD | |
| rs371201709 | 413 | P>T | No |
ESP ExAC gnomAD |
|
| rs1553953917 | 414 | I>V | No | Ensembl | |
| rs781937231 | 416 | V>A | No |
ExAC gnomAD |
|
| rs587627638 | 416 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 416 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1553953954 | 417 | F>L | No | gnomAD | |
| rs1553953954 | 417 | F>V | No | gnomAD | |
| rs147000841 | 418 | L>H | No |
1000Genomes gnomAD |
|
| rs782747449 | 419 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1676541454 | 420 | S>N | No | TOPMed | |
| rs1553953983 | 421 | T>S | No | gnomAD | |
| TCGA novel | 422 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1676544366 TCGA novel |
423 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1450119199 | 423 | A>V | No | TOPMed | |
| rs1553956649 | 424 | G>A | No |
TOPMed gnomAD |
|
| rs1553956649 | 424 | G>D | No |
TOPMed gnomAD |
|
| rs781821502 | 425 | G>R | No |
ExAC gnomAD |
|
| rs1553956680 | 426 | V>A | No |
TOPMed gnomAD |
|
| rs1677967649 | 427 | G>A | No |
TOPMed gnomAD |
|
| rs781902342 | 427 | G>C | No | ExAC | |
| rs1553956694 | 428 | M>I | No |
TOPMed gnomAD |
|
| rs782459357 | 428 | M>T | No |
ExAC gnomAD |
|
| rs1245332567 | 429 | N>D | No | TOPMed | |
| rs1312901526 | 431 | T>A | No | TOPMed | |
| rs201462674 | 432 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs1559830639 | 434 | D>G | No | Ensembl | |
| rs1437131416 | 435 | T>S | No | Ensembl | |
| rs1366064751 | 436 | V>M | No |
TOPMed gnomAD |
|
| rs1677978652 | 438 | F>S | No | TOPMed | |
| rs782224706 | 439 | V>A | No |
ExAC gnomAD |
|
| rs782343138 | 440 | D>G | No |
ExAC gnomAD |
|
| rs781814757 | 441 | S>G | No |
ExAC gnomAD |
|
| rs1319895958 | 441 | S>N | No | Ensembl | |
| rs1677985828 | 444 | N>S | No | Ensembl | |
| rs782423515 | 445 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1677988546 | 446 | Q>* | No | TOPMed | |
| rs1553956777 | 446 | Q>R | No | gnomAD | |
|
rs145656868 COSM895284 |
447 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1553956792 | 447 | N>S | No | gnomAD | |
| rs782062962 | 448 | D>G | No |
ExAC gnomAD |
|
| rs1677994850 | 450 | Q>E | No | Ensembl | |
| rs781963003 | 450 | Q>H | No | ExAC | |
| rs782073787 | 451 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs782073787 | 451 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1678000071 | 453 | A>T | No | TOPMed | |
| TCGA novel | 453 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1678002299 | 454 | R>K | No | gnomAD | |
| TCGA novel | 454 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1367036387 | 455 | A>V | No | TOPMed | |
| COSM3788610 | 456 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1553956866 | 456 | H>R | No |
TOPMed gnomAD |
|
| rs782791630 | 456 | H>Y | No |
ExAC gnomAD |
|
| rs1171065853 | 457 | R>C | No |
TOPMed gnomAD |
|
| rs781853748 | 457 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs746526866 | 458 | I>T | No | Ensembl | |
| rs782164119 | 458 | I>V | No |
ExAC gnomAD |
|
| rs782791468 | 460 | Q>R | No |
ExAC gnomAD |
|
| rs1553956924 | 461 | N>H | No | gnomAD | |
| rs1559831387 | 461 | N>K | No |
TOPMed gnomAD |
|
| rs1678337307 | 462 | K>N | No | TOPMed | |
| rs375312647 | 463 | S>F | No | Ensembl | |
| rs545692282 | 464 | V>A | No | Ensembl | |
|
rs1461398222 COSM5214243 |
466 | V>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs1201399191 | 467 | I>T | No |
TOPMed gnomAD |
|
| rs201774051 | 468 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs587620686 | 468 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs201774051 | 468 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1678348087 | 470 | I>T | No |
TOPMed gnomAD |
|
| rs1678349050 | 471 | G>D | No |
TOPMed gnomAD |
|
| rs1678349050 | 471 | G>V | No |
TOPMed gnomAD |
|
| rs140890542 | 472 | R>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs140890542 | 472 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs200226258 | 472 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1553957637 | 474 | T>A | No | gnomAD | |
| rs1553957643 | 474 | T>I | No | gnomAD | |
| rs1678359420 | 476 | E>K | No | TOPMed | |
| rs372861864 | 477 | E>Q | No |
ESP TOPMed gnomAD |
|
| COSM4021818 | 478 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs143311857 | 478 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs782595117 | 480 | Y>D | No |
ExAC TOPMed gnomAD |
|
| rs782595117 | 480 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1389753499 | 481 | R>K | No |
TOPMed gnomAD |
|
| rs782494783 | 482 | K>I | No |
ExAC gnomAD |
|
| rs1288479963 | 483 | A>P | No |
TOPMed gnomAD |
|
| rs1288479963 | 483 | A>T | No |
TOPMed gnomAD |
|
| rs140343369 | 483 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1678371925 | 484 | A>P | No | TOPMed | |
| rs1370514785 | 485 | S>F | No |
TOPMed gnomAD |
|
| rs1444498992 | 487 | L>R | No | TOPMed | |
| rs1678378222 | 488 | Q>* | No | Ensembl | |
| rs782271333 | 488 | Q>R | No |
ExAC gnomAD |
|
| rs1678381162 | 490 | T>A | No | Ensembl | |
| rs375996768 | 490 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1553957763 | 492 | M>T | No | gnomAD | |
| rs782394217 | 494 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1678384888 | 494 | I>V | No | Ensembl | |
| rs962246846 | 495 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1553957778 | 496 | G>* | No | gnomAD | |
| rs1553957778 | 496 | G>R | No | gnomAD | |
| rs149130696 | 497 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs782167543 | 497 | G>S | No |
ExAC gnomAD |
|
| rs149130696 | 497 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1553957797 | 498 | H>R | No | Ensembl | |
| rs1678395330 | 500 | T>A | No | TOPMed | |
| rs1678397217 | 501 | L>P | No |
TOPMed gnomAD |
|
| rs1678400941 | 502 | G>E | No | TOPMed | |
| rs1553957823 | 502 | G>R | No | gnomAD | |
| rs587773830 | 503 | A>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs920875844 | 503 | A>T | No | gnomAD | |
| rs1223153578 | 504 | Q>* | No | Ensembl | |
|
COSM4932769 rs782743019 COSM4932768 |
504 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs782743019 | 504 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs781817086 | 505 | K>E | No |
ExAC gnomAD |
|
| rs1572000556 | 505 | K>N | No | Ensembl | |
| rs782513568 | 506 | P>A | No |
ExAC gnomAD |
|
| rs1572000642 | 506 | P>L | No | Ensembl | |
| rs367859293 | 507 | A>T | No |
TOPMed gnomAD |
|
| rs1295156078 | 508 | A>T | No |
TOPMed gnomAD |
|
| rs781900246 | 508 | A>V | No |
ExAC gnomAD |
|
| rs781862332 | 509 | D>N | No |
TOPMed gnomAD |
|
| rs1678418713 | 511 | D>E | No | TOPMed | |
| rs587738033 | 512 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs587738033 | 512 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782476079 | 513 | Q>* | No |
ExAC gnomAD |
|
|
COSM4923023 COSM4923022 |
513 | Q>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1410062996 | 515 | S>I | No | TOPMed | |
| rs782014123 | 515 | S>R | No |
ExAC gnomAD |
|
| rs1395205042 | 516 | E>D | No |
TOPMed gnomAD |
|
| rs1680201806 | 517 | I>R | No | Ensembl | |
| COSM4021819 | 517 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782309859 | 518 | L>F | No |
ExAC gnomAD |
|
| rs1680206125 | 519 | K>N | No | TOPMed | |
| rs781952807 | 519 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs782129627 | 520 | F>L | No |
ExAC gnomAD |
|
| rs201567836 | 521 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs587726873 | 522 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781905773 | 522 | L>V | No | ExAC | |
| rs375685901 | 526 | L>M | No |
ESP TOPMed gnomAD |
|
| rs375685901 | 526 | L>V | No |
ESP TOPMed gnomAD |
|
| rs1553960737 | 527 | A>V | No | gnomAD | |
| rs1680216324 | 528 | S>* | No | TOPMed | |
| rs782717581 | 528 | S>F | No | ExAC | |
| rs1680218929 | 529 | E>G | No | TOPMed | |
| rs1553960758 | 530 | G>E | No | gnomAD | |
| rs1553960758 | 530 | G>V | No | gnomAD | |
| rs782651981 | 532 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs782651981 | 532 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1020714011 | 533 | M>I | No |
TOPMed gnomAD |
|
|
rs373424057 RCV000914510 |
533 | M>T | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1553960790 | 534 | D>E | No | gnomAD | |
| rs1553960779 | 534 | D>G | No | gnomAD | |
| rs967797595 | 535 | E>D | No | Ensembl | |
| rs370004935 | 536 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1553960809 | 539 | E>G | No | gnomAD | |
| rs1348881207 | 539 | E>K | No | TOPMed | |
| rs1680230862 | 540 | S>C | No | Ensembl | |
| rs7547279 | 541 | I>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1553960836 | 544 | E>D | No | gnomAD | |
| rs782668430 | 544 | E>Q | No |
ExAC gnomAD |
|
| rs782323069 | 547 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs782323069 | 547 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs782228698 | 548 | G>D | No |
ExAC gnomAD |
|
| rs1553960865 | 549 | Q>* | No | gnomAD | |
| rs1680243027 | 549 | Q>P | No | TOPMed | |
| rs1442168377 | 550 | W>* | No | TOPMed | |
| rs1680246911 | 552 | S>P | No | TOPMed | |
| rs1553960883 | 554 | A>V | No | gnomAD | |
| rs1305657829 | 557 | A>T | No |
TOPMed gnomAD |
|
| rs782423742 | 557 | A>V | No |
ExAC gnomAD |
|
| rs1559853518 | 558 | A>V | No | Ensembl | |
| rs1553960907 | 559 | E>A | No | gnomAD | |
| rs782059967 | 559 | E>D | No | ExAC | |
| rs1680262841 | 560 | G>E | No | gnomAD | |
| rs1375713834 | 560 | G>R | No |
TOPMed gnomAD |
|
| rs1172576878 | 561 | G>E | No |
TOPMed gnomAD |
|
| rs781948152 | 561 | G>W | No |
ExAC gnomAD |
|
| COSM70167 | 562 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782126374 | 563 | R>G | No |
ExAC gnomAD |
|
| rs782749767 | 563 | R>S | No |
ExAC TOPMed gnomAD |
|
|
COSM895286 rs1680272591 COSM4530347 |
564 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1409944475 | 564 | D>V | No | TOPMed | |
| rs587765720 | 565 | Q>E | No |
1000Genomes ExAC gnomAD |
|
| rs1680277019 | 566 | E>Q | No | TOPMed | |
| TCGA novel | 568 | G>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781830629 | 568 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs782302924 | 569 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs781970306 | 570 | N>S | No |
ExAC gnomAD |
|
| rs782078021 | 571 | H>L | No |
ExAC gnomAD |
|
| rs782078021 | 571 | H>R | No |
ExAC gnomAD |
|
| rs1016730630 | 571 | H>Y | No | Ensembl | |
| rs782375872 | 572 | M>I | No |
ExAC gnomAD |
|
| rs1553963839 | 572 | M>R | No | gnomAD | |
| rs1553963839 | 572 | M>T | No | gnomAD | |
| rs1317953767 | 572 | M>V | No | TOPMed | |
| rs782035465 | 574 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1553963851 | 575 | F>L | No |
TOPMed gnomAD |
|
| rs368117603 | 576 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1682201785 | 576 | E>Q | No | gnomAD | |
| rs372796148 | 577 | G>D | No |
ESP TOPMed gnomAD |
|
| rs1682203773 | 577 | G>S | No | TOPMed | |
| rs781795332 | 578 | K>* | No |
ExAC gnomAD |
|
| rs1553963880 | 578 | K>N | No |
TOPMed gnomAD |
|
| rs1553963886 | 579 | D>A | No | gnomAD | |
| rs149791343 | 579 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs149791343 | 579 | D>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs372035697 | 580 | Y>H | No |
ESP TOPMed gnomAD |
|
| rs782729483 | 580 | Y>S | No |
ExAC gnomAD |
|
| rs1388426233 | 581 | S>F | No |
TOPMed gnomAD |
|
| rs373716000 | 582 | K>E | No |
ESP TOPMed gnomAD |
|
| rs1466245306 | 582 | K>N | No |
TOPMed gnomAD |
|
| rs980607852 | 583 | E>G | No | Ensembl | |
| rs1423006272 | 584 | P>S | No |
TOPMed gnomAD |
|
| rs1423006272 | 584 | P>T | No |
TOPMed gnomAD |
|
| rs781879102 | 586 | K>E | No |
ExAC gnomAD |
|
| rs1682220859 | 586 | K>R | No | TOPMed | |
| rs587598833 | 588 | D>E | No |
1000Genomes ExAC gnomAD |
|
| rs927836458 | 588 | D>N | No | gnomAD | |
| TCGA novel | 589 | R>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781882129 | 589 | R>K | No |
ExAC gnomAD |
|
| TCGA novel | 589 | R>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782451462 | 592 | F>S | No |
ExAC gnomAD |
|
| rs1682229176 | 593 | E>G | No | TOPMed | |
|
COSM6121033 COSM6121032 |
593 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781926264 | 594 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1682230372 | 594 | Q>K | No | gnomAD | |
| rs782565499 | 594 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs1270058771 | 595 | L>R | No | Ensembl | |
| rs782340867 | 596 | V>I | No |
ExAC gnomAD |
|
| rs1553963987 | 598 | L>F | No |
TOPMed gnomAD |
|
| rs1553963987 | 598 | L>V | No |
TOPMed gnomAD |
|
| rs1682241041 | 599 | Q>* | No | TOPMed | |
| rs143869639 | 600 | K>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1682244623 | 602 | L>R | No | TOPMed | |
| rs1682243678 | 602 | L>V | No |
TOPMed gnomAD |
|
| rs1682246495 | 604 | E>K | No | gnomAD | |
| rs587665798 | 606 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs782055060 | 607 | S>G | No |
ExAC gnomAD |
|
| rs1682251037 | 608 | Q>* | No | TOPMed | |
| rs1043534922 | 608 | Q>H | No | Ensembl | |
| rs201105160 | 609 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781961050 | 611 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs368838337 | 611 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
|
rs368838337 CA1063844 COSM1730947 RCV000413329 |
611 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA Cosmic ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs781895819 | 612 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs1682255908 | 612 | S>A | No | Ensembl | |
| COSM4021820 | 612 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV000728724 rs587651895 |
614 | R>* | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
COSM2121612 COSM4220189 rs782711426 |
614 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs782486406 | 615 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs781788592 | 615 | N>Y | No |
ExAC TOPMed gnomAD |
|
| rs1329107063 | 617 | G>D | No |
TOPMed gnomAD |
|
| rs1553964636 | 619 | V>A | No | gnomAD | |
| rs1682641236 | 619 | V>I | No | TOPMed | |
| rs1553964639 | 621 | I>M | No | gnomAD | |
| rs2102906165 | 622 | P>A | No | Ensembl | |
| rs1553964649 | 622 | P>L | No |
TOPMed gnomAD |
|
| rs1553964649 | 622 | P>Q | No |
TOPMed gnomAD |
|
| rs1553964649 | 622 | P>R | No |
TOPMed gnomAD |
|
| TCGA novel | 623 | G>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1268723842 | 624 | L>I | No |
TOPMed gnomAD |
|
| TCGA novel | 625 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 627 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1553964676 | 628 | S>T | No |
TOPMed gnomAD |
|
| rs1682649077 | 628 | S>Y | No | Ensembl | |
| rs199838516 | 629 | T>A | No |
TOPMed gnomAD |
|
| rs1553964700 | 630 | K>E | No | gnomAD | |
| rs782474016 | 631 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs373244397 | 632 | K>N | No |
ESP TOPMed gnomAD |
|
| rs1318126587 | 632 | K>R | No |
TOPMed gnomAD |
|
| rs200997862 | 633 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs782545021 | 633 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs200997862 COSM5200386 COSM423716 |
633 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1682658886 | 634 | V>F | No | TOPMed | |
| COSM3473187 | 637 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1553964751 | 642 | D>N | No | gnomAD | |
|
RCV000736171 rs782144677 |
643 | R>missing | No |
ClinVar dbSNP |
|
| rs1392391047 | 643 | R>G | No |
TOPMed gnomAD |
|
| rs782186694 | 644 | Q>* | No |
ExAC gnomAD |
|
| rs782186694 | 644 | Q>E | No |
ExAC gnomAD |
|
| rs376204165 | 645 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs782213864 | 645 | K>R | No |
ExAC gnomAD |
|
| rs782213864 | 645 | K>T | No |
ExAC gnomAD |
|
| rs782391914 | 646 | K>I | No |
ExAC TOPMed |
|
| rs782391914 | 646 | K>R | No |
ExAC TOPMed |
|
| rs782391914 | 646 | K>T | No |
ExAC TOPMed |
|
| rs1682676870 | 647 | R>S | No | TOPMed | |
| rs1682677930 | 648 | Q>* | No | TOPMed | |
| rs2102907530 | 648 | Q>L | No | Ensembl | |
|
rs13374920 VAR_042956 RCV000948556 |
649 | E>A | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs202231038 | 652 | A>G | No |
1000Genomes ExAC gnomAD |
|
|
RCV002227882 rs2102907683 |
652 | A>T | No |
ClinVar Ensembl dbSNP |
|
| rs782707976 | 653 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1553964835 | 655 | R>K | No |
TOPMed gnomAD |
|
| rs782099084 | 656 | R>G | No |
ExAC gnomAD |
|
| rs782748794 | 656 | R>K | No |
ExAC gnomAD |
|
| rs781880621 | 657 | L>F | No |
ExAC gnomAD |
|
| rs1232472154 | 658 | I>K | No |
TOPMed gnomAD |
|
| rs1275162587 | 658 | I>L | No | TOPMed | |
| rs1232472154 | 658 | I>R | No |
TOPMed gnomAD |
|
| rs1232472154 | 658 | I>T | No |
TOPMed gnomAD |
|
| COSM895287 | 658 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782510204 | 659 | E>G | No |
ExAC gnomAD |
|
| rs1682701912 | 659 | E>K | No | Ensembl | |
| rs1553964875 | 660 | E>A | No | gnomAD | |
| rs782817996 | 661 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs142236750 | 662 | K>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs369368487 | 662 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs138412639 | 663 | R>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782647696 | 664 | Q>R | No |
ExAC gnomAD |
|
| rs782246239 | 666 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs782422534 | 667 | E>K | No |
ExAC gnomAD |
|
| rs782586692 | 668 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1382756923 | 669 | E>G | No |
TOPMed gnomAD |
|
| rs1682724350 | 669 | E>K | No | Ensembl | |
| rs1382756923 | 669 | E>V | No |
TOPMed gnomAD |
|
| rs587713818 | 670 | H>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372851796 | 672 | K>R | No |
ESP ExAC gnomAD |
|
| rs1683109073 | 674 | M>I | No |
TOPMed gnomAD |
|
| COSM4021821 | 674 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1559881178 | 674 | M>V | No | Ensembl | |
| rs1553965597 | 675 | A>T | No | gnomAD | |
| rs782699096 | 675 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs781837132 | 676 | W>* | No |
ExAC gnomAD |
|
| rs782470364 | 676 | W>C | No |
ExAC gnomAD |
|
| rs1559881253 | 676 | W>G | No | Ensembl | |
| rs782774749 | 677 | W>C | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 680 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1683115965 | 680 | N>H | No | Ensembl | |
|
rs782558143 RCV000592190 CA1063952 |
680 | N>K | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs201672731 | 680 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs1553965637 | 681 | N>S | No | gnomAD | |
| rs782650323 | 682 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1335439107 | 683 | Q>P | No |
TOPMed gnomAD |
|
| rs1335439107 | 683 | Q>R | No |
TOPMed gnomAD |
|
| rs528197071 | 684 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1553965677 | 685 | F>V | No | gnomAD | |
| rs143929740 | 686 | C>S | No |
ESP TOPMed gnomAD |
|
| rs782218590 | 688 | P>L | No |
ExAC gnomAD |
|
| rs782624893 | 688 | P>S | No |
ExAC gnomAD |
|
| rs148362148 | 691 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1553965731 | 692 | S>G | No | gnomAD | |
| rs587607898 | 692 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1433353778 | 693 | E>A | No |
TOPMed gnomAD |
|
| rs1173564461 | 693 | E>K | No |
TOPMed gnomAD |
|
| rs1553965744 | 694 | P>A | No | gnomAD | |
| rs1553965744 | 694 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| TCGA novel | 695 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1392761683 | 695 | E>K | No | TOPMed | |
| rs1159841268 | 696 | D>E | No |
TOPMed gnomAD |
|
|
COSM3801683 COSM5203423 |
698 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1553965762 | 699 | N>S | No | gnomAD | |
|
rs139791996 RCV002218477 |
700 | G>R | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs782727796 | 703 | S>I | No |
ExAC gnomAD |
|
| rs782001739 | 703 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs782114076 | 704 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs782114076 | 704 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs375815490 | 705 | A>T | No |
ESP ExAC gnomAD |
|
| rs1553965822 | 705 | A>V | No | gnomAD | |
|
rs1443935605 CA342209141 RCV000522058 |
706 | E>G | No |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
| rs587746905 | 706 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs1257896489 | 707 | L>Q | No | TOPMed | |
|
COSM3688970 COSM5134693 |
708 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782720118 | 708 | D>G | No |
ExAC gnomAD |
|
| rs1559882501 | 708 | D>H | No | Ensembl | |
| rs141548764 | 709 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1683148588 | 710 | Q>* | No | Ensembl | |
| rs1224453691 | 710 | Q>R | No |
TOPMed gnomAD |
|
| rs201205825 | 711 | D>Y | No |
1000Genomes gnomAD |
|
| rs1013729045 | 712 | P>Q | No | Ensembl | |
| rs1572138596 | 713 | D>E | No | TOPMed | |
| rs782476225 | 714 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1025544408 | 718 | K>E | No | Ensembl | |
| rs782656610 | 718 | K>R | No | ExAC | |
| rs200761960 | 720 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1559882906 | 721 | S>I | No | Ensembl | |
| rs587733205 | 721 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781953217 | 721 | S>T | No |
ExAC gnomAD |
|
| rs1356531830 | 722 | G>D | No |
TOPMed gnomAD |
|
| rs1683161954 | 723 | D>H | No | Ensembl | |
| rs372787282 | 725 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs372787282 | 725 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs782032017 | 726 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs782318754 | 727 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs782318754 | 727 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs587709474 | 728 | Q>H | No |
1000Genomes ExAC gnomAD |
|
| rs1383533109 | 728 | Q>R | No | Ensembl | |
| rs1553965979 | 730 | G>V | No | gnomAD | |
| rs782105428 | 730 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs1683172388 | 731 | A>S | No | Ensembl | |
| rs782120913 | 732 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs782813032 | 734 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1553966004 | 735 | L>F | No | gnomAD | |
| rs1553966004 | 735 | L>V | No | gnomAD | |
| rs1553966024 | 736 | I>T | No | gnomAD | |
| rs148675923 | 738 | H>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782625612 | 738 | H>R | No | ExAC | |
| rs148675923 | 738 | H>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs142193146 | 740 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs142193146 COSM1472546 COSM5229854 |
740 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC TOPMed gnomAD NCI-TCGA Cosmic NCI-TCGA |
| rs782402608 | 741 | D>H | No | ExAC | |
| rs1683723539 | 742 | D>G | No | gnomAD | |
| rs782610593 | 742 | D>Y | No |
ExAC gnomAD |
|
| rs782223724 | 743 | S>A | No |
ExAC gnomAD |
|
|
VAR_042957 RCV001674491 rs2275249 |
743 | S>C | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2275249 | 743 | S>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2102944320 | 744 | G>A | No | Ensembl | |
| rs1683729637 | 744 | G>S | No | TOPMed | |
| rs12033160 | 745 | H>Q | No |
TOPMed gnomAD |
|
| rs781854060 | 745 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs782630219 | 745 | H>Y | No |
ExAC gnomAD |
|
| rs1553967058 | 746 | W>* | No | gnomAD | |
| rs1683735309 | 746 | W>R | No | Ensembl | |
| rs1045551687 | 747 | G>D | No |
TOPMed gnomAD |
|
| rs1045551687 | 747 | G>V | No |
TOPMed gnomAD |
|
| rs1683739459 | 749 | G>S | No | TOPMed | |
| rs782404802 | 751 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs781935687 | 752 | F>L | No |
ExAC gnomAD |
|
| rs1358593430 | 752 | F>L | No |
TOPMed gnomAD |
|
| rs1553967091 | 753 | T>R | No |
TOPMed gnomAD |
|
| rs1174245880 | 756 | E>Q | No | TOPMed | |
| rs1683747069 | 757 | K>E | No | Ensembl | |
| rs190301424 | 758 | R>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs370710748 | 758 | R>L | No |
ExAC TOPMed gnomAD |
|
|
COSM5228107 COSM423718 |
758 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370710748 | 758 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs782817356 | 759 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1683751086 | 759 | S>P | No | gnomAD | |
| rs782687350 | 760 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs782687350 | 760 | A>S | No |
ExAC TOPMed gnomAD |
|
| COSM1688474 | 760 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1683756472 | 761 | E>* | No | Ensembl | |
| rs1683757358 | 762 | P>A | No |
TOPMed gnomAD |
|
| rs1260311857 | 762 | P>R | No |
TOPMed gnomAD |
|
| rs1683757358 | 762 | P>S | No |
TOPMed gnomAD |
|
| rs375162292 | 763 | R>G | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 763 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1022311001 | 764 | K>E | No | Ensembl | |
| rs1683761019 | 764 | K>R | No | TOPMed | |
|
RCV002114114 rs148289715 |
765 | I>M | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1254865890 | 765 | I>T | No |
TOPMed gnomAD |
|
| rs781832900 | 765 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs587597741 | 766 | Y>C | No |
1000Genomes ExAC gnomAD |
|
| rs1553967171 | 766 | Y>N | No | gnomAD | |
| rs781859702 | 768 | L>V | No |
ExAC gnomAD |
|
| rs986647106 | 769 | A>D | No | Ensembl | |
| rs1553967196 | 769 | A>S | No | gnomAD | |
| rs782550003 | 770 | G>A | No |
ExAC gnomAD |
|
| rs782550003 | 770 | G>E | No |
ExAC gnomAD |
|
| rs1348328085 | 770 | G>R | No |
TOPMed gnomAD |
|
| rs1348328085 | 770 | G>W | No |
TOPMed gnomAD |
|
| rs1683774733 | 771 | K>E | No | Ensembl | |
| rs143800773 | 771 | K>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs143800773 | 771 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs782450061 | 772 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs782431108 | 772 | M>R | No | Ensembl | |
| TCGA novel | 773 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1683781024 | 773 | K>Q | No | Ensembl | |
| rs1398737020 | 774 | D>E | No |
TOPMed gnomAD |
|
| rs201898934 | 774 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1685509984 | 775 | L>P | No | Ensembl | |
| rs782292994 | 776 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs782292994 | 776 | S>G | No |
ExAC TOPMed gnomAD |
|
|
rs61756300 RCV000969409 |
776 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD ClinVar dbSNP |
|
| rs1452160882 | 777 | L>F | No |
TOPMed gnomAD |
|
| rs1553970441 | 779 | G>A | No | gnomAD | |
| rs1553970441 | 779 | G>D | No | gnomAD | |
| rs868931664 | 779 | G>S | No | gnomAD | |
| rs1179017911 | 780 | V>A | No |
TOPMed gnomAD |
|
| rs1685519282 | 780 | V>I | No | Ensembl | |
| rs76912870 | 782 | L>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs782054927 | 783 | F>I | No |
ExAC TOPMed gnomAD |
|
| rs782054927 | 783 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1443036471 | 785 | V>I | No |
TOPMed gnomAD |
|
| rs1310544526 | 788 | K>* | No | TOPMed | |
| rs782587273 | 791 | R>G | No |
ExAC gnomAD |
|
| rs782266228 | 792 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs2103005527 | 793 | K>N | No | Ensembl | |
| TCGA novel | 793 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1375353471 | 794 | G>V | No |
TOPMed gnomAD |
|
| rs1286425690 | 795 | Q>E | No | TOPMed | |
| rs1449356648 | 796 | D>A | No |
TOPMed gnomAD |
|
| rs1553970560 | 796 | D>H | No | TOPMed | |
| rs1449356648 | 796 | D>V | No |
TOPMed gnomAD |
|
|
RCV000523146 rs1553970560 CA342211124 |
796 | D>Y | No |
ClinGen ClinVar TOPMed dbSNP |
|
| rs1420386811 | 798 | L>S | No |
TOPMed gnomAD |
|
| rs150536851 | 799 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs781804099 | 800 | L>S | No |
ExAC TOPMed gnomAD |
|
| rs1553972931 | 801 | I>S | No |
TOPMed gnomAD |
|
| rs1553972931 | 801 | I>T | No |
TOPMed gnomAD |
|
| rs1686427991 | 802 | V>L | No | TOPMed | |
| rs1259425316 | 803 | A>T | No |
TOPMed gnomAD |
|
|
RCV000911849 rs149664186 RCV000594735 CA1064099 |
804 | Q>* | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs782298619 | 806 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs147666732 | 806 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs781837246 | 807 | D>H | No |
TOPMed gnomAD |
|
|
rs782680914 COSM3730124 COSM4220212 |
808 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1006441553 | 808 | R>H | No |
TOPMed gnomAD |
|
| rs1553973054 | 809 | S>C | No | gnomAD | |
| rs1553973045 | 809 | S>P | No | gnomAD | |
| rs782199063 | 810 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs782361440 | 811 | V>I | No |
ExAC gnomAD |
|
| TCGA novel | 812 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1686443845 | 812 | L>P | No | Ensembl | |
| rs782111228 | 813 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs1553973096 | 814 | G>R | No | gnomAD | |
| rs1553973102 | 814 | G>V | No | gnomAD | |
| rs1553973106 | 816 | K>N | No |
TOPMed gnomAD |
|
| rs1400464743 | 817 | M>I | No | gnomAD | |
| rs1294338469 | 817 | M>K | No |
TOPMed gnomAD |
|
| rs1294338469 | 817 | M>T | No |
TOPMed gnomAD |
|
| rs782014875 | 818 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1553973148 | 818 | A>V | No |
TOPMed gnomAD |
|
| rs145436064 | 820 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs140537285 | 821 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs140537285 | 821 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1471434499 | 823 | G>D | No | TOPMed | |
| rs1157757508 | 823 | G>S | No |
TOPMed gnomAD |
|
| rs1419532152 | 824 | L>P | No |
TOPMed gnomAD |
|
| rs782077111 | 825 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1686468369 | 825 | K>R | No | TOPMed | |
| rs2103032331 | 826 | K>E | No | Ensembl | |
| rs1559917841 | 827 | I>K | No | Ensembl | |
|
rs148434783 RCV002175135 |
827 | I>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs782570086 | 828 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA |
| rs1423054249 | 828 | F>L | No |
TOPMed gnomAD |
|
| TCGA novel | 829 | L>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1253751844 | 830 | A>G | No |
TOPMed gnomAD |
|
| rs1253751844 | 830 | A>V | No |
TOPMed gnomAD |
|
| rs201165419 | 831 | A>P | No |
1000Genomes ExAC gnomAD |
|
| rs1553973253 | 831 | A>V | No |
TOPMed gnomAD |
|
| rs782015121 | 833 | K>E | No | Ensembl | |
| rs1553974340 | 837 | S>T | No | gnomAD | |
| TCGA novel | 838 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369192147 | 838 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1026015295 | 839 | H>R | No |
TOPMed gnomAD |
|
| rs2103041406 | 839 | H>Y | No | Ensembl | |
| rs587766046 | 840 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs112733685 | 840 | L>P | No | Ensembl | |
| rs1553974416 | 841 | P>A | No | gnomAD | |
| rs2103041519 | 841 | P>L | No | Ensembl | |
|
COSM674766 rs1553974416 |
841 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| rs782175604 | 842 | R>C | No |
ExAC TOPMed gnomAD |
|
|
VAR_086613 rs782473109 COSM4021822 |
842 | R>H | Variant assessed as Somatic; MODERATE impact. found in patients with cancer; loss of auto-inhibition, leading to constitutive ATP-dependent chromatin remodeler activity [NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic UniProt ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs782595154 | 843 | I>S | No |
ExAC gnomAD |
|
| rs782595154 | 843 | I>T | No |
ExAC gnomAD |
|
| rs782260077 | 844 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1553974480 | 845 | H>R | No |
TOPMed gnomAD |
|
| rs782369690 | 845 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs868984459 | 846 | A>D | No | Ensembl | |
| rs1553974486 | 846 | A>T | No | gnomAD | |
| rs868984459 | 846 | A>V | No | Ensembl | |
| rs373116065 | 847 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs148225662 | 848 | K>E | No |
ESP TOPMed gnomAD |
|
| rs587718472 | 849 | G>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs587718472 | 849 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1358136615 | 849 | G>V | No | TOPMed | |
|
COSM423719 rs1553974573 VAR_086614 COSM5832265 |
852 | W>C | Variant assessed as Somatic; MODERATE impact. found in patients with cancer; loss of auto-inhibition, leading to constitutive ATP-dependent chromatin remodeler activity [NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD UniProt dbSNP |
| rs1173615115 | 853 | Y>C | No |
TOPMed gnomAD |
|
| rs782723083 | 854 | G>V | No |
ExAC gnomAD |
|
| rs1572256773 | 855 | T>I | No | Ensembl | |
| rs1572256773 | 855 | T>S | No | Ensembl | |
| rs2103042169 | 856 | E>Q | No | Ensembl | |
| rs782007432 | 857 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
VAR_086615 COSM3399673 rs1180954265 |
857 | R>Q | Variant assessed as Somatic; MODERATE impact. found in patients with cancer; loss of auto-inhibition, leading to constitutive ATP-dependent chromatin remodeler activity [NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic UniProt NCI-TCGA TOPMed dbSNP |
| TCGA novel | 858 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782120249 | 859 | I>N | No |
ExAC gnomAD |
|
| rs782120249 | 859 | I>T | No |
ExAC gnomAD |
|
| rs139712616 | 860 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs377751265 | 860 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs139712616 VAR_086616 COSM895289 |
860 | R>W | Variant assessed as Somatic; MODERATE impact. found in patients with cancer; loss of auto-inhibition, leading to constitutive ATP-dependent chromatin remodeler activity [NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic UniProt ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs782759083 | 862 | H>R | No |
ExAC gnomAD |
|
| rs2103042431 | 864 | A>P | No | Ensembl | |
| TCGA novel | 864 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781816199 | 864 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2103042483 | 865 | A>T | No | Ensembl | |
| rs1553974704 | 866 | R>G | No |
TOPMed gnomAD |
|
| rs1686840683 | 867 | G>D | No | gnomAD | |
| rs782521400 | 867 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1686840683 | 867 | G>V | No | gnomAD | |
| rs2103042604 | 868 | I>M | No | Ensembl | |
| rs1553974729 | 868 | I>T | No | gnomAD | |
| rs782630838 | 869 | P>R | No |
ExAC gnomAD |
|
| COSM4021823 | 870 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1553974755 | 870 | T>I | No |
TOPMed gnomAD |
|
| rs782497810 | 871 | Y>* | No |
ExAC gnomAD |
|
| rs2103042704 | 871 | Y>N | No | Ensembl | |
| rs370660814 | 872 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs782541065 | 872 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs781901195 | 872 | I>V | No |
ExAC gnomAD |
|
| rs1182316367 | 873 | Y>H | No |
TOPMed gnomAD |
|
| rs1553976151 | 876 | P>H | No | gnomAD | |
| rs1553976151 | 876 | P>L | No | gnomAD | |
| rs1687187927 | 877 | R>K | No | gnomAD | |
| rs201834043 | 878 | S>G | No |
1000Genomes gnomAD |
|
| rs782231915 | 878 | S>N | No |
ExAC gnomAD |
|
| rs1687190827 | 879 | K>* | No | Ensembl | |
| rs587765141 | 880 | S>F | No |
1000Genomes ExAC gnomAD |
|
| rs587765141 | 880 | S>Y | No |
1000Genomes ExAC gnomAD |
|
| rs1553976206 | 881 | A>T | No | gnomAD | |
| rs1241349424 | 882 | V>I | No | TOPMed | |
| rs1241349424 | 882 | V>L | No | TOPMed | |
| rs1465492698 | 883 | L>F | No |
TOPMed gnomAD |
|
| rs1553976228 | 884 | H>N | No | gnomAD | |
| rs1687199325 | 884 | H>P | No | TOPMed | |
|
rs4950394 RCV000952812 VAR_042958 |
885 | S>A | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs4950394 | 885 | S>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs4950394 | 885 | S>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs782103548 | 886 | Q>K | No |
ExAC gnomAD |
|
| rs782429928 | 886 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs782429928 | 886 | Q>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 886 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1323833951 | 891 | S>F | No |
TOPMed gnomAD |
|
| rs782010267 | 892 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs782010267 | 892 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs1381492597 | 893 | R>G | No | TOPMed | |
| rs1553976361 | 894 | Q>* | No |
TOPMed gnomAD |
|
| rs1553976361 | 894 | Q>E | No |
TOPMed gnomAD |
|
| rs201349327 | 896 | V>L | No |
ExAC TOPMed gnomAD |
|
|
RCV000994093 rs201349327 |
896 | V>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1687216552 | 897 | P>A | No | TOPMed | |
| rs782092066 | 897 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1687218148 | 898 | P>E | No | TOPMed |
No associated diseases with Q86WJ1
6 regional properties for Q86WJ1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SNF2, N-terminal | 49 - 327 | IPR000330 |
| domain | Helicase, C-terminal domain-like | 348 - 513 | IPR001650 |
| conserved_site | DNA/RNA helicase, ATP-dependent, DEAH-box type, conserved site | 169 - 178 | IPR002464 |
| domain | Macro domain | 704 - 897 | IPR002589 |
| domain | Helicase superfamily 1/2, ATP-binding domain | 42 - 230 | IPR014001 |
| domain | SNF2/RAD5-like, C-terminal helicase domain | 346 - 470 | IPR049730 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.- | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47157 | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1-LIKE |
| PANTHER Subfamily | PTHR47157:SF1 | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1-LIKE |
| PANTHER Protein Class | chromatin/chromatin-binding, or -regulatory protein | |
| PANTHER Pathway Category |
Wnt signaling pathway SWI/SNF |
|
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| site of DNA damage | A region of a chromosome at which DNA damage has occurred. DNA damage signaling and repair proteins accumulate at the lesion to respond to the damage and repair the DNA to form a continuous DNA helix. |
| site of double-strand break | A region of a chromosome at which a DNA double-strand break has occurred. DNA damage signaling and repair proteins accumulate at the lesion to respond to the damage and repair the DNA to form a continuous DNA helix. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| ATP-dependent chromatin remodeler activity | An activity, driven by ATP hydrolysis, that modulates the contacts between histones and DNA, resulting in a change in chromosome architecture within the nucleosomal array, leading to chromatin remodeling. |
| DNA helicase activity | Unwinding of a DNA helix, driven by ATP hydrolysis. |
| histone reader activity | A chromatin adaptor that recognizes specific forms of histones, either modified by a post-translational modification, or the unmodified form. Histone readers have roles in many processes, including in centromere function or in modulating the accessibility of cis-regulatory regions to the transcription machinery. |
| nucleosome binding | Binding to a nucleosome, a complex comprised of DNA wound around a multisubunit core and associated proteins, which forms the primary packing unit of DNA into higher order structures. |
| nucleotide binding | Binding to a nucleotide, any compound consisting of a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the ribose or deoxyribose. |
| poly-ADP-D-ribose modification-dependent protein binding | Binding to a protein upon poly-ADP-ribosylation of the target protein. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| chromatin remodeling | A dynamic process of chromatin reorganization resulting in changes to chromatin structure. These changes allow DNA metabolic processes such as transcriptional regulation, DNA recombination, DNA repair, and DNA replication. |
| DNA damage response | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| DNA repair | The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3B7N1 | CHD1L | Chromodomain-helicase-DNA-binding protein 1-like | Bos taurus (Bovine) | SS |
| Q24368 | Iswi | Chromatin-remodeling complex ATPase chain Iswi | Drosophila melanogaster (Fruit fly) | SS |
| O60264 | SMARCA5 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 | Homo sapiens (Human) | EV |
| P28370 | SMARCA1 | Probable global transcription activator SNF2L1 | Homo sapiens (Human) | SS |
| Q9NRZ9 | HELLS | Lymphoid-specific helicase | Homo sapiens (Human) | PR |
| Q60848 | Hells | Lymphocyte-specific helicase | Mus musculus (Mouse) | PR |
| Q91ZW3 | Smarca5 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 | Mus musculus (Mouse) | SS |
| Q6PGB8 | Smarca1 | Probable global transcription activator SNF2L1 | Mus musculus (Mouse) | SS |
| Q9CXF7 | Chd1l | Chromodomain-helicase-DNA-binding protein 1-like | Mus musculus (Mouse) | SS |
| Q7G8Y3 | Os01g0367900 | Probable chromatin-remodeling complex ATPase chain | Oryza sativa subsp japonica (Rice) | PR |
| P41877 | isw-1 | Chromatin-remodeling complex ATPase chain isw-1 | Caenorhabditis elegans | SS |
| Q8RWY3 | CHR11 | ISWI chromatin-remodeling complex ATPase CHR11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9XFH4 | DDM1 | ATP-dependent DNA helicase DDM1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MERAGATSRG | GQAPGFLLRL | HTEGRAEAAR | VQEQDLRQWG | LTGIHLRSYQ | LEGVNWLAQR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FHCQNGCILG | DEMGLGKTCQ | TIALFIYLAG | RLNDEGPFLI | LCPLSVLSNW | KEEMQRFAPG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LSCVTYAGDK | EERACLQQDL | KQESRFHVLL | TTYEICLKDA | SFLKSFPWSV | LVVDEAHRLK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NQSSLLHKTL | SEFSVVFSLL | LTGTPIQNSL | QELYSLLSFV | EPDLFSKEEV | GDFIQRYQDI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EKESESASEL | HKLLQPFLLR | RVKAEVATEL | PKKTEVVIYH | GMSALQKKYY | KAILMKDLDA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| FENETAKKVK | LQNILSQLRK | CVDHPYLFDG | VEPEPFEVGD | HLTEASGKLH | LLDKLLAFLY |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SGGHRVLLFS | QMTQMLDILQ | DYMDYRGYSY | ERVDGSVRGE | ERHLAIKNFG | QQPIFVFLLS |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TRAGGVGMNL | TAADTVIFVD | SDFNPQNDLQ | AAARAHRIGQ | NKSVKVIRLI | GRDTVEEIVY |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RKAASKLQLT | NMIIEGGHFT | LGAQKPAADA | DLQLSEILKF | GLDKLLASEG | STMDEIDLES |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ILGETKDGQW | VSDALPAAEG | GSRDQEEGKN | HMYLFEGKDY | SKEPSKEDRK | SFEQLVNLQK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| TLLEKASQEG | RSLRNKGSVL | IPGLVEGSTK | RKRVLSPEEL | EDRQKKRQEA | AAKRRRLIEE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| KKRQKEEAEH | KKKMAWWESN | NYQSFCLPSE | ESEPEDLENG | EESSAELDYQ | DPDATSLKYV |
| 730 | 740 | 750 | 760 | 770 | 780 |
| SGDVTHPQAG | AEDALIVHCV | DDSGHWGRGG | LFTALEKRSA | EPRKIYELAG | KMKDLSLGGV |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LLFPVDDKES | RNKGQDLLAL | IVAQHRDRSN | VLSGIKMAAL | EEGLKKIFLA | AKKKKASVHL |
| 850 | 860 | 870 | 880 | 890 | |
| PRIGHATKGF | NWYGTERLIR | KHLAARGIPT | YIYYFPRSKS | AVLHSQSSSS | SSRQLVP |