Q86VH2
Gene name |
KIF27 |
Protein name |
Kinesin-like protein KIF27 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:55582 |
EC number |
|
Protein Class |
|
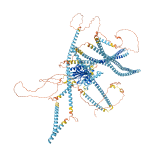
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
0-575 (N-terminal motor domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q86VH2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q86VH2-F1 | Predicted | AlphaFoldDB |
1415 variants for Q86VH2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs528345101 | 4 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs773009927 | 4 | I>T | No |
ExAC gnomAD |
|
| rs528345101 | 4 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1463890071 | 5 | P>L | No | TOPMed | |
| rs1466260647 | 9 | A>V | No |
TOPMed gnomAD |
|
| rs748128352 | 10 | V>I | No |
ExAC gnomAD |
|
| rs1409619237 | 12 | I>T | No | gnomAD | |
| rs1268097096 | 12 | I>V | No | TOPMed | |
| rs1435102395 | 13 | R>G | No | gnomAD | |
| rs746054884 | 14 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs746054884 | 14 | P>S | No |
ExAC TOPMed gnomAD |
|
| COSM6183819 | 17 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779259691 | 17 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs779259691 | 17 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs544957472 | 18 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1955600678 | 19 | E>K | No |
TOPMed gnomAD |
|
| rs1955600144 | 20 | A>S | No | Ensembl | |
| rs1955600144 | 20 | A>T | No | Ensembl | |
| rs1390587633 | 22 | H>D | No | TOPMed | |
| rs1955599342 | 22 | H>Q | No | Ensembl | |
| rs151072016 | 23 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1313623453 | 24 | H>N | No | gnomAD | |
| rs1955598440 | 24 | H>R | No | Ensembl | |
| rs1955598003 | 25 | Q>P | No | TOPMed | |
| rs201835321 | 27 | C>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1375126193 | 28 | V>M | No | gnomAD | |
| rs759936405 | 30 | V>L | No | ExAC | |
| rs377450779 | 31 | I>V | No | Ensembl | |
| rs1448128443 | 33 | N>D | No | gnomAD | |
| rs1040967379 | 33 | N>S | No |
TOPMed gnomAD |
|
| rs773990415 | 34 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1344406739 | 35 | Q>* | No |
TOPMed gnomAD |
|
| rs1467611843 | 36 | Q>R | No |
TOPMed gnomAD |
|
| rs1378442265 | 38 | I>V | No | gnomAD | |
| rs1955593908 | 39 | I>T | No | TOPMed | |
| rs1270362653 | 41 | R>K | No |
TOPMed gnomAD |
|
| rs762527132 | 41 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs761818271 | 42 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs772737449 | 42 | D>G | No |
ExAC gnomAD |
|
| rs772737449 | 42 | D>V | No |
ExAC gnomAD |
|
| rs768564047 | 44 | V>G | No | ExAC | |
| rs776747895 | 44 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1458179490 | 46 | T>N | No | gnomAD | |
| rs1285172100 | 48 | D>H | No |
TOPMed gnomAD |
|
| rs1227834005 | 49 | F>S | No | gnomAD | |
| rs1955591244 | 52 | G>D | No | TOPMed | |
| rs1955590750 | 56 | T>A | No | gnomAD | |
| rs746856988 | 57 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1256282163 | 58 | D>G | No | gnomAD | |
| rs1413859659 | 59 | E>D | No | TOPMed | |
| rs779723498 | 59 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1352007772 | 60 | V>A | No | gnomAD | |
| rs1471517746 | 62 | N>K | No | TOPMed | |
| rs749506941 | 64 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs541660256 | 64 | C>R | No |
1000Genomes ExAC gnomAD |
|
| rs749506941 | 64 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs148934176 | 65 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs148934176 | 65 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1955587749 | 66 | K>E | No | Ensembl | |
| rs756237758 | 66 | K>M | No |
ExAC gnomAD |
|
| rs1397921850 | 67 | P>S | No | gnomAD | |
| rs1955586687 | 68 | L>R | No | Ensembl | |
| rs781715428 | 68 | L>V | No |
ExAC gnomAD |
|
| rs370476698 | 69 | V>G | No |
ESP TOPMed gnomAD |
|
| rs1955586019 | 70 | L>V | No | TOPMed | |
| rs755324153 | 71 | S>T | No |
ExAC gnomAD |
|
| rs751893763 | 73 | I>T | No |
ExAC gnomAD |
|
| rs1349001779 | 75 | G>S | No | Ensembl | |
| rs1564018176 | 75 | G>V | No | Ensembl | |
| rs555892893 | 76 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs766658858 | 76 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs750091863 | 78 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs184212344 | 80 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs184212344 | 80 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1487116477 | 81 | F>L | No | gnomAD | |
| rs1287667036 | 83 | Y>C | No |
TOPMed gnomAD |
|
| TCGA novel | 85 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1207529635 | 86 | T>I | No |
TOPMed gnomAD |
|
| rs1207529635 | 86 | T>S | No |
TOPMed gnomAD |
|
| rs1955581585 | 89 | G>E | No | gnomAD | |
| TCGA novel | 93 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768654011 | 94 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs760689665 | 94 | I>T | No |
ExAC gnomAD |
|
| rs768654011 | 94 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs145300186 | 95 | G>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs771788991 | 97 | G>V | No | ExAC | |
| rs1398188008 | 98 | H>R | No |
TOPMed gnomAD |
|
| rs1362700694 | 99 | I>S | No | gnomAD | |
| rs777900219 | 100 | A>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777900219 | 100 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs769948226 | 100 | A>V | No |
ExAC gnomAD |
|
| rs776652629 | 102 | V>I | No |
ExAC gnomAD |
|
| rs1244585494 | 103 | V>G | No |
TOPMed gnomAD |
|
| TCGA novel | 104 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1469143368 | 104 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1954821796 | 104 | E>Q | No | Ensembl | |
| rs747519922 | 105 | G>A | No |
ExAC gnomAD |
|
| rs1320142314 | 108 | G>C | No |
TOPMed gnomAD |
|
| rs1954819472 | 108 | G>D | No |
TOPMed gnomAD |
|
| rs1320142314 | 108 | G>S | No |
TOPMed gnomAD |
|
| rs139715507 | 109 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1293994664 | 110 | I>V | No |
TOPMed gnomAD |
|
| rs147431079 | 112 | R>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs147431079 | 112 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1035562287 | 112 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1276762658 | 113 | A>G | No |
TOPMed gnomAD |
|
| rs1438361984 | 114 | I>V | No | gnomAD | |
| rs567251057 | 115 | Q>* | No |
1000Genomes ExAC gnomAD |
|
| rs753563611 | 115 | Q>R | No |
ExAC gnomAD |
|
| rs1954816813 | 117 | I>M | No | Ensembl | |
| rs763679267 | 118 | F>L | No |
ExAC gnomAD |
|
| rs763679267 | 118 | F>V | No |
ExAC gnomAD |
|
| rs755624703 | 119 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs752678080 | 119 | Q>R | No |
ExAC gnomAD |
|
| rs1422641795 | 120 | S>N | No | gnomAD | |
| rs1173957621 | 122 | S>Y | No |
TOPMed gnomAD |
|
| rs1005104667 | 123 | E>D | No | TOPMed | |
| rs1564003861 | 124 | H>Q | No |
TOPMed gnomAD |
|
| rs759532647 | 124 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs774067614 | 125 | P>R | No |
ExAC gnomAD |
|
| rs1954813360 | 125 | P>S | No | TOPMed | |
| rs1954810429 | 126 | S>G | No | Ensembl | |
| rs766193831 | 126 | S>N | No |
ExAC gnomAD |
|
| rs761975954 | 128 | D>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 133 | V>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 133 | V>Y | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775649967 | 134 | S>C | No |
ExAC gnomAD |
|
| rs772389559 | 135 | Y>C | No |
ExAC gnomAD |
|
| rs772389559 | 135 | Y>F | No |
ExAC gnomAD |
|
| rs532273915 | 135 | Y>H | No | Ensembl | |
| rs1237641815 | 136 | I>L | No | gnomAD | |
| rs746414083 | 136 | I>M | No |
ExAC gnomAD |
|
| rs1263300643 | 137 | E>Q | No |
TOPMed gnomAD |
|
| rs375810143 | 138 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs779367580 | 138 | V>M | No |
ExAC gnomAD |
|
| rs1335821601 | 139 | Y>* | No | gnomAD | |
| rs1356928913 | 139 | Y>C | No |
TOPMed gnomAD |
|
| rs748921730 | 140 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1359442310 | 141 | E>G | No | gnomAD | |
| rs755640552 | 142 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs777637489 | 142 | D>Y | No |
ExAC gnomAD |
|
| rs767060247 | 145 | D>E | No |
ExAC gnomAD |
|
| rs1239264304 | 146 | L>F | No | gnomAD | |
| rs1258227049 | 146 | L>R | No |
TOPMed gnomAD |
|
| rs1954803607 | 148 | E>Q | No | Ensembl | |
| rs372555424 | 151 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs372555424 | 151 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
|
rs766130139 COSM1110631 |
153 | M>I | endometrium Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated ExAC TOPMed gnomAD NCI-TCGA Cosmic |
| rs140491572 | 153 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs140491572 | 153 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1318494334 | 154 | K>R | No | gnomAD | |
| rs762802842 | 155 | D>G | No |
ExAC gnomAD |
|
| rs2132673364 | 156 | L>V | No | Ensembl | |
| rs1168950204 | 157 | H>P | No | TOPMed | |
| rs1168950204 | 157 | H>R | No | TOPMed | |
| rs1376399526 | 158 | I>N | No | gnomAD | |
| rs1227640125 | 158 | I>V | No | gnomAD | |
|
COSM1110630 rs537684585 |
159 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1954799028 | 159 | R>Q | No |
TOPMed gnomAD |
|
| rs1379335676 | 160 | E>Q | No |
TOPMed gnomAD |
|
| rs772212322 | 161 | D>E | No |
ExAC gnomAD |
|
| rs923880280 | 162 | E>D | No | gnomAD | |
| rs569974082 | 162 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1954797243 | 163 | K>Q | No | TOPMed | |
| rs1346152563 | 163 | K>R | No | gnomAD | |
| TCGA novel | 164 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775041607 | 165 | N>D | No |
ExAC gnomAD |
|
| rs200513825 | 166 | T>A | No | gnomAD | |
| rs200513825 | 166 | T>S | No | gnomAD | |
| rs1229629138 | 167 | V>M | No |
TOPMed gnomAD |
|
| rs1437555116 | 169 | V>A | No | TOPMed | |
| rs771510121 | 169 | V>F | No |
ExAC gnomAD |
|
| rs763559372 | 170 | G>E | No |
ExAC gnomAD |
|
| rs540462841 | 172 | K>Q | No |
1000Genomes ExAC gnomAD |
|
| rs765323788 | 173 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1048536036 | 175 | H>L | No |
TOPMed gnomAD |
|
| rs1255077081 | 176 | V>A | No |
TOPMed gnomAD |
|
| rs1217896307 | 177 | E>* | No |
TOPMed gnomAD |
|
| rs1217896307 | 177 | E>Q | No |
TOPMed gnomAD |
|
| rs199846865 | 178 | S>G | No |
1000Genomes ExAC gnomAD |
|
| rs1954218857 | 178 | S>R | No | TOPMed | |
| rs1310130356 | 179 | A>T | No | Ensembl | |
| rs1954217868 | 180 | G>D | No | TOPMed | |
| rs780963076 | 181 | E>K | No |
ExAC gnomAD |
|
| rs1333478872 | 183 | M>I | No | gnomAD | |
| rs2132590195 | 183 | M>R | No | Ensembl | |
| rs1954216167 | 184 | S>N | No | Ensembl | |
|
rs746463879 COSM3927129 |
185 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs746463879 | 185 | L>I | No |
ExAC gnomAD |
|
| TCGA novel | 187 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1196816 | 188 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1563992947 | 188 | M>R | No | Ensembl | |
| rs1954214804 | 189 | G>E | No | gnomAD | |
| rs1954214124 | 190 | N>K | No | gnomAD | |
| rs147985902 | 191 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs758289085 | 194 | H>R | No | ExAC | |
| rs1954213383 | 194 | H>Y | No |
TOPMed gnomAD |
|
| rs369710588 | 195 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1954212304 | 197 | T>A | No | Ensembl | |
| rs1954211940 | 197 | T>N | No | Ensembl | |
| rs1396626977 | 198 | T>P | No | gnomAD | |
| rs921525533 | 199 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs778547102 | 201 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs757112864 | 202 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1954210047 | 202 | E>K | No | Ensembl | |
| rs752861336 | 203 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs752861336 | 203 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs965411515 | 203 | H>R | No | TOPMed | |
| rs767839569 | 205 | S>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 206 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM95622 rs755267943 |
207 | S>* | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1440944907 | 208 | H>Q | No |
TOPMed gnomAD |
|
| rs1207322796 | 209 | A>E | No | gnomAD | |
| rs144421081 | 210 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5103335 COSM3764033 RCV000455189 VAR_035361 rs12001918 |
213 | I>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1392092015 | 215 | I>V | No | gnomAD | |
| rs1301357262 | 216 | C>F | No | gnomAD | |
| rs763505930 | 218 | V>A | No |
ExAC gnomAD |
|
| rs1376251326 | 221 | N>S | No | gnomAD | |
| rs1376251326 | 221 | N>T | No | gnomAD | |
| rs1438791398 | 222 | M>I | No | gnomAD | |
| rs771235706 | 222 | M>T | No |
ExAC gnomAD |
|
| rs765488881 | 224 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1954201850 | 224 | A>V | No |
TOPMed gnomAD |
|
| rs1419196338 | 228 | G>R | No | gnomAD | |
| rs1954200403 | 230 | W>* | No | Ensembl | |
| rs1305114587 | 230 | W>R | No | gnomAD | |
| rs1954200065 | 231 | Y>C | No | gnomAD | |
| rs1198185761 | 232 | S>C | No |
TOPMed gnomAD |
|
| rs1198185761 | 232 | S>F | No |
TOPMed gnomAD |
|
| rs776870488 | 232 | S>T | No |
ExAC gnomAD |
|
| rs1954198890 | 233 | P>R | No | TOPMed | |
| rs1280512260 | 234 | R>L | No | gnomAD | |
| rs1280512260 | 234 | R>Q | No | gnomAD | |
| rs747324827 | 234 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1954197066 | 236 | I>V | No | TOPMed | |
| rs958131422 | 238 | S>* | No | TOPMed | |
| rs1954195736 | 240 | F>Y | No | TOPMed | |
| rs1236887444 | 241 | H>Y | No | gnomAD | |
| rs1954194644 | 243 | V>A | No | Ensembl | |
| rs774927647 | 244 | D>E | No |
ExAC gnomAD |
|
| rs1230232525 | 244 | D>H | No | gnomAD | |
| rs1230232525 | 244 | D>N | No | gnomAD | |
| rs372426841 | 247 | G>V | No |
TOPMed gnomAD |
|
| rs149773175 | 253 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141363838 | 254 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs141363838 | 254 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs141363838 | 254 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs777437927 | 255 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1954189609 | 256 | N>S | No | TOPMed | |
| rs1954189299 | 257 | T>A | No | TOPMed | |
| rs1179124812 | 258 | G>A | No | Ensembl | |
| rs1954188695 | 259 | E>Q | No | Ensembl | |
| rs751853807 | 260 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs139609439 COSM4696119 |
260 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
COSM94583 rs193285297 |
261 | F>L | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1014123547 | 262 | K>E | No | TOPMed | |
| rs138255319 | 262 | K>R | No |
ESP TOPMed gnomAD |
|
|
rs1954186553 COSM1110629 |
263 | E>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs1954186185 | 264 | S>C | No | Ensembl | |
|
COSM1674579 rs1382476055 |
266 | Q>R | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1954185486 | 267 | I>V | No | Ensembl | |
| rs376710039 | 268 | N>D | No |
ESP TOPMed gnomAD |
|
| rs1160279911 | 268 | N>S | No |
TOPMed gnomAD |
|
| rs758591940 | 271 | L>F | No |
ExAC gnomAD |
|
| rs750879422 | 273 | A>D | No |
ExAC gnomAD |
|
| rs1403012309 | 273 | A>P | No | TOPMed | |
| TCGA novel | 273 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1432578382 | 274 | L>F | No | gnomAD | |
| rs765715657 | 274 | L>S | No |
ExAC gnomAD |
|
| rs1475867389 | 275 | G>* | No |
TOPMed gnomAD |
|
| TCGA novel | 277 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1192473790 | 278 | I>M | No | gnomAD | |
| rs762230637 | 278 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs777012575 | 279 | S>N | No |
ExAC gnomAD |
|
| rs372449530 | 280 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1450519156 | 280 | A>V | No | Ensembl | |
| rs775160703 | 281 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs140475602 | 283 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs771563536 | 283 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs771563536 | 283 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs151248208 | 284 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
| rs151248208 | 284 | P>R | No |
ESP TOPMed gnomAD |
|
|
COSM3848981 rs1277851962 |
285 | R>C | breast [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs773750084 | 285 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs770839077 | 286 | R>G | No |
ExAC gnomAD |
|
| rs1954176471 | 287 | K>R | No | Ensembl | |
| rs1439310727 | 288 | S>N | No | gnomAD | |
| COSM3908546 | 289 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1954175175 | 290 | H>R | No | TOPMed | |
| rs1954175524 | 290 | H>Y | No | Ensembl | |
| rs150593704 | 293 | Y>C | No |
1000Genomes ExAC gnomAD |
|
| rs777663766 | 296 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs187494265 | 296 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs747204801 | 298 | I>T | No |
ExAC gnomAD |
|
|
rs35594736 VAR_035362 |
300 | R>Q | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs368536781 | 300 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1563991276 | 301 | L>I | No | Ensembl | |
| rs750448963 | 301 | L>P | No |
ExAC gnomAD |
|
| rs1194365160 | 302 | L>V | No | gnomAD | |
| rs1451382418 | 304 | D>N | No | gnomAD | |
| rs374690692 | 305 | S>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs374690692 | 305 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1110628 rs374690692 |
305 | S>Y | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC TOPMed gnomAD |
| rs142339049 | 306 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs142339049 | 306 | L>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1254876416 | 306 | L>V | No | TOPMed | |
| TCGA novel | 307 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1469005121 | 310 | A>T | No | gnomAD | |
| rs1257531814 | 310 | A>V | No | gnomAD | |
| rs754346636 | 311 | K>E | No |
ExAC gnomAD |
|
| rs548369061 | 311 | K>N | No |
1000Genomes ExAC gnomAD |
|
| rs1341746414 | 311 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs147859809 | 313 | V>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147859809 | 313 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147859809 | 313 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs773875283 | 314 | M>I | No |
ExAC gnomAD |
|
| rs759119585 | 314 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs113210971 | 316 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1407657054 | 317 | C>R | No |
TOPMed gnomAD |
|
| COSM3908545 | 318 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1168134045 | 319 | S>I | No |
TOPMed gnomAD |
|
| rs1168134045 | 319 | S>N | No |
TOPMed gnomAD |
|
| rs116477156 | 321 | S>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs532390425 | 321 | S>T | No |
1000Genomes ExAC gnomAD |
|
| rs1954159007 | 322 | S>F | No | Ensembl | |
| rs746349752 | 323 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs781137516 | 323 | S>T | No |
ExAC gnomAD |
|
| rs746349752 | 323 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs779025482 | 325 | F>S | No |
ExAC gnomAD |
|
| rs1245602994 | 328 | S>P | No | Ensembl | |
| TCGA novel | 329 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs754337057 COSM3782304 |
331 | S>C | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs754337057 | 331 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs754337057 | 331 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs1954155586 | 332 | L>V | No | TOPMed | |
| rs778307249 | 333 | K>R | No |
ExAC gnomAD |
|
| rs144003628 | 334 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs144003628 | 334 | Y>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1277553362 | 335 | A>V | No |
TOPMed gnomAD |
|
| rs767312772 | 336 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs751221009 | 339 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs759182637 | 339 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs762439007 | 340 | N>S | No |
ExAC gnomAD |
|
| rs1193133819 | 341 | I>M | No |
TOPMed gnomAD |
|
| rs773271183 | 341 | I>V | No |
ExAC gnomAD |
|
| rs769785786 | 344 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1954150799 | 345 | P>T | No | TOPMed | |
| rs1454938551 | 346 | T>I | No |
TOPMed gnomAD |
|
| rs776517224 | 348 | N>T | No |
ExAC gnomAD |
|
| rs1954149589 | 349 | F>L | No | TOPMed | |
| rs1233501644 | 350 | S>I | No |
TOPMed gnomAD |
|
| TCGA novel | 352 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777838110 | 352 | E>D | No |
ExAC gnomAD |
|
|
COSM1255843 rs146210501 |
352 | E>K | Variant assessed as Somatic; MODERATE impact. oesophagus [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs146210501 | 352 | E>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1563990156 | 353 | S>L | No | Ensembl | |
| TCGA novel | 354 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756577075 | 355 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs748681327 | 355 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs756577075 | 355 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs751530679 | 356 | I>T | No |
TOPMed gnomAD |
|
| rs781598275 | 356 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs755339000 | 357 | D>E | No |
ExAC gnomAD |
|
| rs1014153336 | 357 | D>G | No |
TOPMed gnomAD |
|
| rs1331665191 | 357 | D>N | No |
TOPMed gnomAD |
|
| rs1954144150 | 359 | M>I | No | TOPMed | |
| rs1353198773 | 362 | E>G | No | gnomAD | |
| TCGA novel | 362 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766007800 | 365 | L>F | No |
ExAC gnomAD |
|
|
COSM257239 rs371692315 |
367 | R>* | Variant assessed as Somatic; HIGH impact. kidney large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs367896750 | 367 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs764861313 | 368 | E>K | No |
ExAC gnomAD |
|
| rs764861313 | 368 | E>Q | No |
ExAC gnomAD |
|
| rs761820172 | 369 | A>P | No |
ExAC gnomAD |
|
| rs761820172 | 369 | A>T | No |
ExAC gnomAD |
|
| rs763941203 | 370 | L>S | No |
ExAC gnomAD |
|
| rs1563989819 | 372 | S>G | No | Ensembl | |
| rs1346490413 | 372 | S>N | No | TOPMed | |
| rs1362206149 | 372 | S>R | No |
TOPMed gnomAD |
|
| rs1262081344 | 373 | Q>R | No |
TOPMed gnomAD |
|
| TCGA novel | 374 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1954139359 | 375 | A>T | No | Ensembl | |
| rs1485905562 | 376 | G>D | No | gnomAD | |
| rs200024207 | 376 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200024207 | 376 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770993949 | 378 | S>N | No |
ExAC gnomAD |
|
| rs1954137427 | 379 | Q>* | No | TOPMed | |
| rs368483166 | 379 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1204977675 | 379 | Q>R | No | TOPMed | |
| rs1318666012 | 380 | T>P | No | gnomAD | |
| rs1318666012 | 380 | T>S | No | gnomAD | |
| rs1456000826 | 381 | T>A | No |
TOPMed gnomAD |
|
| rs1284800216 | 381 | T>I | No |
TOPMed gnomAD |
|
| rs1284800216 | 381 | T>N | No |
TOPMed gnomAD |
|
| rs1954135261 | 382 | Q>H | No | Ensembl | |
| rs41282417 | 383 | I>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs866702256 | 384 | N>I | No | TOPMed | |
| rs866702256 | 384 | N>S | No | TOPMed | |
|
COSM1674578 rs1414992383 |
385 | R>* | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs888073995 | 385 | R>L | No |
TOPMed gnomAD |
|
| rs888073995 | 385 | R>Q | No |
TOPMed gnomAD |
|
| rs1954132822 | 386 | E>D | No | Ensembl | |
| rs1362219300 | 387 | G>E | No |
TOPMed gnomAD |
|
| rs139709598 | 388 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1486938889 | 389 | P>S | No | gnomAD | |
|
rs1954130856 COSM1110627 |
390 | D>A | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs1954130856 | 390 | D>G | No | TOPMed | |
| rs1346320705 | 390 | D>H | No | gnomAD | |
| rs1164686382 | 391 | T>P | No | gnomAD | |
| TCGA novel | 392 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1459979876 | 393 | R>G | No | gnomAD | |
| rs755358229 | 393 | R>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 393 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1954128676 | 394 | I>M | No | TOPMed | |
| rs780496824 | 394 | I>V | No |
ExAC gnomAD |
|
| rs758073293 | 397 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs780768715 | 398 | E>G | No | Ensembl | |
| rs1184578875 | 399 | E>K | No | gnomAD | |
| rs1486719827 | 400 | Q>R | No | gnomAD | |
| rs756890196 | 405 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1306564074 | 406 | G>E | No | gnomAD | |
| rs753797148 | 406 | G>R | No |
ExAC gnomAD |
|
| rs1306564074 | 406 | G>V | No | gnomAD | |
| rs1954124683 | 407 | E>D | No | TOPMed | |
| rs996995862 | 410 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1216749502 | 411 | Y>H | No |
TOPMed gnomAD |
|
| rs763878405 | 413 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs199704500 | 415 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 417 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs192546948 | 418 | A>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs192546948 | 418 | A>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs192546948 | 418 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2132573538 | 419 | F>I | No | Ensembl | |
| rs138516740 | 420 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs138516740 | 420 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| COSM2154217 | 423 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776787113 | 423 | V>G | No |
ExAC gnomAD |
|
| TCGA novel | 424 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1179085000 | 426 | K>R | No |
TOPMed gnomAD |
|
| rs1954119787 | 427 | D>H | No | Ensembl | |
| rs200613968 | 427 | D>V | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 429 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780259075 | 429 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs780259075 | 429 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs569987566 | 432 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs370980782 | 432 | N>T | No |
ESP TOPMed gnomAD |
|
|
COSM3433308 rs77150451 |
433 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1954116799 | 435 | Q>* | No | Ensembl | |
| rs778762556 | 435 | Q>R | No |
ExAC gnomAD |
|
| rs1954115680 | 437 | H>N | No | Ensembl | |
| rs1954115409 | 438 | K>* | No |
TOPMed gnomAD |
|
| rs776832186 | 441 | E>Q | No | Ensembl | |
| rs376468427 | 442 | W>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1299427057 | 443 | F>L | No |
TOPMed gnomAD |
|
| rs1312939713 | 445 | M>I | No | TOPMed | |
| rs1232467396 | 445 | M>T | No |
TOPMed gnomAD |
|
| rs915767148 | 445 | M>V | No |
TOPMed gnomAD |
|
| rs755958827 | 446 | I>F | No |
ExAC gnomAD |
|
| rs992636851 | 446 | I>M | No | TOPMed | |
| rs1954111988 | 447 | Q>L | No | Ensembl | |
| rs759394387 | 450 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs750631509 | 451 | K>N | No |
ExAC gnomAD |
|
| rs765387138 | 452 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs765387138 | 452 | A>T | No |
ExAC TOPMed gnomAD |
|
| COSM3848980 | 456 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3927128 rs1954108690 |
456 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1954108952 | 456 | S>P | No | TOPMed | |
|
COSM1110626 rs1194182598 |
458 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs768828645 | 458 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs148142223 | 461 | G>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs148142223 | 461 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs772548524 | 462 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1480756080 | 462 | G>S | No | gnomAD | |
| rs1476071541 | 463 | T>P | No |
TOPMed gnomAD |
|
| rs1476071541 | 463 | T>S | No |
TOPMed gnomAD |
|
| rs1350492713 | 464 | A>P | No | gnomAD | |
| rs1416860069 | 466 | L>M | No | TOPMed | |
| rs1238939275 | 468 | E>K | No |
TOPMed gnomAD |
|
| COSM487616 | 469 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1954103394 | 470 | P>S | No | TOPMed | |
| rs779421731 | 471 | Q>R | No |
ExAC gnomAD |
|
| rs748995650 | 472 | H>R | No |
ExAC gnomAD |
|
| rs770669830 | 472 | H>Y | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs1954101481 |
473 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1563988025 | 474 | T>I | No | Ensembl | |
| rs1311277947 | 477 | Q>* | No | gnomAD | |
| rs147375672 | 477 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1954099223 | 479 | K>R | No | gnomAD | |
| rs142728940 | 481 | E>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1412535917 | 482 | L>F | No | TOPMed | |
| rs1954097406 | 483 | K>R | No | TOPMed | |
| rs983063564 | 484 | K>T | No |
TOPMed gnomAD |
|
| rs1954096848 | 485 | C>W | No | TOPMed | |
| rs1293101889 | 486 | Q>E | No | TOPMed | |
| rs1183665947 | 487 | C>Y | No |
TOPMed gnomAD |
|
| rs903207904 | 489 | L>R | No | TOPMed | |
| rs769178563 | 491 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1953669210 | 492 | D>G | No | Ensembl | |
| rs1192940923 | 492 | D>N | No | gnomAD | |
| rs149402364 | 493 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1953668536 | 494 | V>A | No | Ensembl | |
| COSM261666 | 494 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1953668210 | 495 | V>L | No | gnomAD | |
| rs1953667882 | 496 | F>L | No | Ensembl | |
| rs780687564 | 496 | F>Y | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 497 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1953667213 | 498 | Q>R | No | TOPMed | |
| rs1953666497 | 500 | E>* | No | Ensembl | |
| rs754873051 | 501 | L>M | No |
ExAC gnomAD |
|
| rs746994795 | 503 | V>L | No |
ExAC gnomAD |
|
| rs746994795 | 503 | V>M | No |
ExAC gnomAD |
|
| rs758276554 | 505 | E>K | No |
ExAC gnomAD |
|
| rs1953663652 | 506 | L>P | No |
TOPMed gnomAD |
|
| rs138849280 | 507 | K>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1335084537 | 507 | K>N | No |
TOPMed gnomAD |
|
| rs1953661959 | 509 | Q>R | No | Ensembl | |
| rs1588239843 | 512 | M>I | No | Ensembl | |
| COSM3908544 | 512 | M>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756294201 | 512 | M>V | No |
ExAC gnomAD |
|
| rs1953660881 | 513 | M>V | No | TOPMed | |
| rs1953660182 | 515 | Q>E | No | gnomAD | |
| rs767725843 | 516 | E>* | No |
ExAC gnomAD |
|
| rs371540279 | 518 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1244724762 | 519 | G>A | No |
TOPMed gnomAD |
|
| rs1166960257 | 521 | A>D | No | gnomAD | |
| COSM1490182 | 522 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1341837683 | 523 | S>F | No | TOPMed | |
| TCGA novel | 526 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs550945409 | 527 | A>G | No |
1000Genomes ExAC gnomAD |
|
| rs550945409 | 527 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs1454224976 | 528 | Q>E | No | gnomAD | |
| TCGA novel | 532 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs147660786 | 532 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs746941840 | 532 | R>S | No |
ExAC gnomAD |
|
| rs766736854 | 534 | Q>P | No |
TOPMed gnomAD |
|
| rs766736854 | 534 | Q>R | No |
TOPMed gnomAD |
|
| COSM3659211 | 536 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1308254432 | 537 | K>I | No | gnomAD | |
|
COSM282237 rs1414592449 |
537 | K>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1374000797 | 538 | I>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1952675747 | 538 | I>V | No | TOPMed | |
| TCGA novel | 539 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1110624 | 540 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1192030341 | 541 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs770892449 | 541 | Q>R | No |
ExAC gnomAD |
|
| rs867090787 | 542 | Q>R | No | Ensembl | |
| rs1952674298 | 543 | L>F | No | TOPMed | |
| rs1407813175 | 543 | L>H | No |
TOPMed gnomAD |
|
| rs371440040 | 544 | L>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs371440040 | 544 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs371839444 | 544 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1952672731 | 546 | D>V | No | gnomAD | |
| rs1252899633 | 549 | S>G | No |
TOPMed gnomAD |
|
| rs1252899633 | 549 | S>R | No |
TOPMed gnomAD |
|
| rs751626818 | 553 | T>R | No |
ExAC TOPMed gnomAD |
|
| COSM257238 | 554 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1719306 | 555 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs183482711 | 559 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1323979320 | 560 | T>A | No |
TOPMed gnomAD |
|
| rs1563964477 | 562 | S>* | No | TOPMed | |
| rs1952668154 | 563 | A>T | No | Ensembl | |
| rs1952667644 | 565 | E>D | No | TOPMed | |
| rs749127015 | 566 | N>S | No | Ensembl | |
| rs762219846 | 567 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs762219846 | 567 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1288640763 | 569 | D>H | No | gnomAD | |
| TCGA novel | 569 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 570 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1952665491 | 570 | G>R | No |
TOPMed gnomAD |
|
| rs1952664933 | 571 | P>L | No | TOPMed | |
| rs1952664621 | 572 | D>V | No | gnomAD | |
| COSM3908543 | 573 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs368013775 | 574 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs775520025 | 578 | R>K | No |
TOPMed gnomAD |
|
| rs1952663126 | 579 | R>G | No | TOPMed | |
| rs1281188082 | 580 | P>T | No | TOPMed | |
| rs771529286 | 581 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1952661784 | 582 | T>A | No | TOPMed | |
| rs2132373394 | 583 | V>A | No | Ensembl | |
| rs759487755 | 583 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs759487755 | 583 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1474367536 | 584 | P>Q | No |
TOPMed gnomAD |
|
| rs1484624833 | 584 | P>S | No | gnomAD | |
| rs749034071 | 585 | F>C | No |
ExAC TOPMed gnomAD |
|
| rs1481234693 | 585 | F>L | No | Ensembl | |
| rs749034071 | 585 | F>S | No |
ExAC TOPMed gnomAD |
|
| COSM4831147 | 586 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3659210 rs1952657954 |
588 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| COSM1463307 | 589 | L>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1952657671 | 590 | G>R | No | Ensembl | |
| rs1196828858 | 590 | G>V | No | TOPMed | |
| rs1952656801 | 591 | H>R | No | Ensembl | |
| rs2132372630 | 593 | I>V | No | Ensembl | |
| rs1563963892 | 594 | Y>C | No | Ensembl | |
| rs780163512 | 595 | I>M | No |
ExAC gnomAD |
|
| rs140861753 | 595 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2132372390 | 597 | S>L | No | Ensembl | |
| rs537046371 | 598 | R>K | No |
1000Genomes ExAC gnomAD |
|
| COSM6116003 | 598 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1161925764 | 599 | Q>K | No | TOPMed | |
| rs1212447189 | 600 | D>H | No |
TOPMed gnomAD |
|
| rs1362531361 | 600 | D>V | No |
TOPMed gnomAD |
|
| TCGA novel | 600 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750925868 | 603 | K>R | No |
ExAC gnomAD |
|
| rs1168896026 | 605 | H>D | No | TOPMed | |
| rs1273240986 | 605 | H>R | No |
TOPMed gnomAD |
|
| rs370389309 | 607 | S>G | No |
ESP TOPMed gnomAD |
|
| rs150984973 | 608 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 608 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201326496 | 608 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1207821907 | 609 | P>L | No | gnomAD | |
| rs1207821907 | 609 | P>R | No | gnomAD | |
| TCGA novel | 609 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1232764485 | 610 | M>R | No |
TOPMed gnomAD |
|
| rs1232764485 | 610 | M>T | No |
TOPMed gnomAD |
|
| rs776485584 | 610 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs560511993 | 611 | Y>* | No |
1000Genomes TOPMed |
|
| rs142641392 | 612 | S>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1332245616 | 612 | S>P | No | TOPMed | |
| rs774392277 | 614 | D>N | No |
ExAC gnomAD |
|
| rs774392277 | 614 | D>Y | No |
ExAC gnomAD |
|
| rs1371528379 | 615 | R>* | No |
TOPMed gnomAD |
|
| rs1310085211 | 615 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1952430538 | 615 | R>SI* | No | Ensembl | |
| rs574858708 | 616 | I>M | No |
1000Genomes ExAC gnomAD |
|
| rs762558596 | 616 | I>V | No | Ensembl | |
| rs1319665719 | 618 | A>S | No | TOPMed | |
| rs1952429150 | 619 | G>A | No | gnomAD | |
| rs1952429150 | 619 | G>E | No | gnomAD | |
| rs749789850 | 619 | G>R | No |
ExAC gnomAD |
|
|
COSM3659209 rs148451065 COSM3659208 |
621 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs375590529 COSM1463306 |
621 | R>Q | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1952428176 | 622 | T>A | No | gnomAD | |
| rs1379231698 | 622 | T>I | No | gnomAD | |
| rs748394033 | 623 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs748394033 | 623 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM1212450 COSM3908542 rs367944774 |
623 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA gnomAD |
| rs754542614 | 624 | S>C | No |
ExAC gnomAD |
|
| rs993037050 | 624 | S>N | No |
TOPMed gnomAD |
|
| rs1483150342 | 626 | M>L | No |
TOPMed gnomAD |
|
| rs544396672 | 626 | M>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs758038163 | 627 | L>M | No |
ExAC gnomAD |
|
| rs749964270 | 627 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1193440438 | 629 | G>D | No | Ensembl | |
| rs765013499 | 631 | I>V | No |
ExAC TOPMed gnomAD |
|
|
COSM6183823 COSM6183824 |
634 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753527642 | 634 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs753527642 | 634 | Q>R | No |
ExAC TOPMed gnomAD |
|
|
COSM4927919 rs763998301 COSM4927920 |
636 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1952422708 | 637 | V>D | No | Ensembl | |
| rs1952422960 | 637 | V>I | No | Ensembl | |
| rs577155251 | 638 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs577155251 | 638 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1952421974 | 639 | H>Y | No |
TOPMed gnomAD |
|
| rs1952420588 | 640 | C>* | No | Ensembl | |
| rs1170958659 | 640 | C>R | No | Ensembl | |
| rs1401563486 | 640 | C>Y | No |
TOPMed gnomAD |
|
| rs1015883192 | 641 | Q>R | No |
TOPMed gnomAD |
|
| rs1404341011 | 644 | D>G | No |
TOPMed gnomAD |
|
| rs770987050 | 645 | N>S | No |
ExAC gnomAD |
|
| rs773284705 | 646 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs773284705 | 646 | S>R | No |
ExAC TOPMed gnomAD |
|
|
COSM3659206 COSM3659205 |
648 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs187689772 | 648 | D>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1160683485 | 648 | D>H | No |
TOPMed gnomAD |
|
| rs1160683485 | 648 | D>N | No |
TOPMed gnomAD |
|
|
RCV000926431 rs187689772 |
648 | D>V | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1160683485 | 648 | D>Y | No |
TOPMed gnomAD |
|
| rs1416635822 | 650 | E>Q | No | gnomAD | |
| rs769106774 | 651 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs146702826 | 651 | S>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146702826 | 651 | S>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146702826 | 651 | S>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1952416548 | 653 | G>D | No | Ensembl | |
| rs746658775 | 654 | Q>K | No |
ExAC gnomAD |
|
| rs1952415694 | 654 | Q>R | No | TOPMed | |
|
COSM1110622 COSM4869676 |
655 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1191407851 | 655 | E>K | No |
TOPMed gnomAD |
|
| rs1287671817 | 656 | K>E | No | TOPMed | |
| rs1487527130 | 657 | S>Y | No | TOPMed | |
|
CA5103037 RCV000454440 rs13289566 |
658 | G>E | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1952413882 | 659 | T>A | No | TOPMed | |
| rs758126112 | 660 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs771798020 | 660 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1270129058 | 660 | R>T | No | gnomAD | |
| rs1363012307 | 661 | C>S | No |
TOPMed gnomAD |
|
| rs1952344336 | 661 | C>W | No | Ensembl | |
| rs745564312 | 661 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs190657428 | 662 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM754007 COSM4860180 |
662 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756856861 | 664 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs535421391 | 664 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs777710144 | 665 | S>P | No |
ExAC gnomAD |
|
| rs756051676 | 667 | I>L | No |
ExAC gnomAD |
|
|
COSM6116005 COSM6116004 |
667 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM456164 COSM4815742 |
668 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1379133426 | 670 | P>R | No |
TOPMed gnomAD |
|
| rs139819631 | 672 | S>C | No |
ESP ExAC gnomAD |
|
| rs762013627 | 679 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs201164159 | 683 | Q>R | No | 1000Genomes | |
| rs570677869 | 684 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1195075544 | 686 | T>A | No |
TOPMed gnomAD |
|
| rs1195075544 | 686 | T>P | No |
TOPMed gnomAD |
|
| rs762929444 | 687 | Q>L | No |
ExAC gnomAD |
|
| rs762929444 | 687 | Q>R | No |
ExAC gnomAD |
|
| rs772604037 | 690 | D>Y | No |
ExAC gnomAD |
|
| rs1347014094 | 691 | L>S | No |
TOPMed gnomAD |
|
|
COSM6116007 COSM6116006 |
692 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1272580868 | 693 | N>K | No | gnomAD | |
| rs1952336959 | 693 | N>S | No | Ensembl | |
| rs745511203 | 695 | D>N | No |
ExAC gnomAD |
|
| rs1268178135 | 698 | I>M | No | TOPMed | |
| rs1156987912 | 698 | I>V | No | gnomAD | |
| rs781168308 | 699 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1473700057 | 699 | D>H | No |
TOPMed gnomAD |
|
| rs768597394 | 701 | L>F | No |
ExAC gnomAD |
|
| rs779731823 | 702 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs1442891004 | 702 | Q>H | No | gnomAD | |
| rs951236153 | 705 | Q>R | No |
TOPMed gnomAD |
|
|
COSM3848979 COSM1110621 |
706 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1330844231 | 707 | L>* | No | gnomAD | |
| rs554432970 | 708 | N>D | No |
1000Genomes ExAC gnomAD |
|
| rs1212670856 | 709 | L>F | No |
TOPMed gnomAD |
|
| rs756317975 | 710 | Q>E | No |
ExAC gnomAD |
|
| rs1486137261 | 711 | K>Q | No | gnomAD | |
| rs1449445659 | 712 | L>* | No |
TOPMed gnomAD |
|
| rs2132299319 | 712 | L>I | No | Ensembl | |
| rs1318640583 | 713 | K>N | No | gnomAD | |
| rs752836636 | 713 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs867801714 | 715 | S>* | No | Ensembl | |
|
COSM1331466 rs138059115 |
717 | R>C | ovary [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs766704130 | 717 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs766704130 | 717 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs138059115 | 717 | R>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1404604246 | 718 | I>V | No | gnomAD | |
| rs541364399 | 720 | T>P | No |
1000Genomes ExAC gnomAD |
|
| rs772918709 | 722 | A>T | No |
ExAC gnomAD |
|
| rs769282175 | 725 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 726 | M>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM6116009 COSM6116008 |
726 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1463305 COSM5100795 |
726 | M>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1247329101 | 726 | M>T | No | gnomAD | |
|
COSM1331467 rs543373060 |
727 | R>T | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs868641474 | 728 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs868641474 | 728 | E>K | No |
TOPMed gnomAD |
|
| rs1315186909 | 730 | T>I | No |
TOPMed gnomAD |
|
| rs1315186909 | 730 | T>K | No |
TOPMed gnomAD |
|
| rs776146281 | 731 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs746966836 | 733 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs746966836 | 733 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1415739171 | 734 | K>T | No |
TOPMed gnomAD |
|
| rs139434047 | 735 | M>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139434047 | 735 | M>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs749510445 | 736 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs150902888 | 738 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1296041458 | 739 | L>M | No |
TOPMed gnomAD |
|
| rs1952172059 | 739 | L>R | No |
TOPMed gnomAD |
|
| rs1296041458 | 739 | L>V | No |
TOPMed gnomAD |
|
| rs1365995874 | 744 | I>M | No |
TOPMed gnomAD |
|
| rs2132297747 | 744 | I>T | No | Ensembl | |
| rs752785370 | 745 | K>Q | No |
ExAC gnomAD |
|
|
COSM3908537 COSM3908536 |
747 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772057381 | 748 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs202151820 | 749 | D>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs774317958 | 750 | A>V | No |
ExAC gnomAD |
|
| rs1588155591 | 751 | K>E | No | Ensembl | |
| rs1951885472 | 751 | K>N | No | Ensembl | |
| rs1951885734 | 751 | K>T | No | Ensembl | |
| rs1422398093 | 753 | V>I | No |
TOPMed gnomAD |
|
| rs1951884013 | 755 | K>R | No |
TOPMed gnomAD |
|
| rs189628967 | 756 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1430950474 | 758 | S>F | No | gnomAD | |
| rs781303597 | 758 | S>P | No |
ExAC gnomAD |
|
| rs1482497079 | 760 | K>Q | No |
TOPMed gnomAD |
|
| rs747540513 | 761 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1214002721 | 762 | T>A | No | gnomAD | |
| rs1317996326 | 764 | L>P | No | TOPMed | |
| rs1951881278 | 764 | L>V | No | Ensembl | |
| rs1951880160 | 765 | E>D | No | TOPMed | |
| rs368787925 | 765 | E>G | No |
ESP ExAC gnomAD |
|
| rs879897580 | 768 | A>S | No |
TOPMed gnomAD |
|
| rs879897580 | 768 | A>T | No |
TOPMed gnomAD |
|
| rs758953455 | 769 | E>G | No |
ExAC TOPMed gnomAD |
|
|
COSM4834680 COSM4834681 |
769 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1255917235 | 771 | A>G | No | gnomAD | |
| rs750733298 | 772 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1313999918 | 773 | V>A | No | gnomAD | |
| rs756868923 | 774 | E>* | No |
ExAC TOPMed gnomAD |
|
|
COSM3848978 rs756868923 COSM1212446 |
774 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs763645542 | 776 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1286243481 | 776 | I>T | No |
TOPMed gnomAD |
|
| rs760044042 | 777 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs551413413 | 778 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs551413413 | 778 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767316258 | 781 | Q>H | No |
ExAC gnomAD |
|
| rs1564368205 | 781 | Q>K | No | Ensembl | |
| rs1951875829 | 783 | Q>P | No | TOPMed | |
| rs2132251973 | 787 | N>D | No | Ensembl | |
| rs774266514 | 788 | K>E | No |
ExAC gnomAD |
|
| rs770892402 | 788 | K>R | No |
ExAC gnomAD |
|
| rs748377938 | 789 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs532983093 | 790 | L>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780216959 | 795 | M>I | No | ExAC | |
| rs746997899 | 795 | M>T | No |
ExAC gnomAD |
|
| rs374771696 | 795 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs746410542 | 797 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs746410542 | 797 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs779242004 | 799 | L>F | No |
ExAC TOPMed gnomAD |
|
|
COSM4870082 COSM1110620 |
800 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1951870526 | 802 | E>* | No | TOPMed | |
|
COSM1212447 rs754214393 |
804 | R>C | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs371848145 | 804 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs371848145 | 804 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs754214393 | 804 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs752099024 | 805 | K>E | No |
ExAC gnomAD |
|
| rs767105041 | 805 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs759515743 | 806 | K>E | No |
ExAC gnomAD |
|
| rs759515743 | 806 | K>Q | No |
ExAC gnomAD |
|
| rs946063233 | 807 | M>I | No |
TOPMed gnomAD |
|
| rs774499624 | 808 | D>G | No |
ExAC gnomAD |
|
| rs774499624 | 808 | D>V | No |
ExAC gnomAD |
|
| rs1951867885 | 809 | A>T | No | TOPMed | |
| rs1951867633 | 810 | A>T | No | gnomAD | |
| rs1951867374 | 810 | A>V | No | Ensembl | |
| rs540980899 | 811 | K>E | No |
1000Genomes ExAC gnomAD |
|
| rs879022136 | 812 | L>V | No |
TOPMed gnomAD |
|
|
COSM4862386 COSM754008 |
813 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1160891774 | 813 | R>S | No |
TOPMed gnomAD |
|
| rs1054433149 | 815 | Q>E | No |
TOPMed gnomAD |
|
| rs755700582 | 816 | V>G | No |
ExAC gnomAD |
|
| rs200139828 | 818 | Q>* | No |
TOPMed gnomAD |
|
| rs200139828 | 818 | Q>E | No |
TOPMed gnomAD |
|
| rs537443766 | 820 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs754596155 | 820 | K>R | No |
ExAC gnomAD |
|
| rs12686150 | 821 | Q>H | No | Ensembl | |
| TCGA novel | 822 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766356232 | 822 | Q>L | No |
ExAC gnomAD |
|
| rs763019549 | 826 | K>N | No |
ExAC gnomAD |
|
| rs1951583856 | 826 | K>T | No | Ensembl | |
| rs376225572 | 829 | S>L | No |
ESP TOPMed gnomAD |
|
| rs1414364725 | 830 | L>V | No | TOPMed | |
| rs1223017955 | 831 | S>P | No | gnomAD | |
|
COSM1110619 COSM4863823 |
832 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765116309 | 833 | Q>H | No |
ExAC gnomAD |
|
| rs760799438 | 834 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs772293053 | 835 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs371881517 | 837 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1362596650 | 837 | R>H | No | TOPMed | |
| rs1398221802 | 838 | A>T | No |
TOPMed gnomAD |
|
| rs1951580653 | 838 | A>V | No | Ensembl | |
| TCGA novel | 840 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1110618 COSM4869577 |
842 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771377650 | 845 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1436957339 | 846 | D>E | No | TOPMed | |
| rs777969757 | 848 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs777969757 | 848 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs556438812 | 849 | K>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1168631219 | 850 | Y>* | No | gnomAD | |
| rs568433898 | 850 | Y>H | No | Ensembl | |
| rs1477720009 | 851 | Q>* | No | gnomAD | |
| rs1477720009 | 851 | Q>E | No | gnomAD | |
| rs1327319732 | 851 | Q>R | No | gnomAD | |
| rs1951576317 | 852 | K>T | No | TOPMed | |
| rs1951575821 | 853 | I>V | No | TOPMed | |
| rs1269337789 | 854 | Q>E | No | gnomAD | |
|
COSM3908534 COSM3908535 |
855 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs946769129 | 855 | L>P | No | TOPMed | |
| rs754684305 | 857 | R>T | No |
ExAC gnomAD |
|
| rs1564361742 | 858 | K>* | No | Ensembl | |
| rs538049404 | 858 | K>N | No |
1000Genomes ExAC |
|
|
rs779736930 COSM5998273 COSM487614 |
860 | R>* | Variant assessed as Somatic; HIGH impact. kidney [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs200952027 | 860 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs761608802 | 861 | E>D | No |
ExAC gnomAD |
|
| rs570508641 | 861 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs1173294339 | 862 | E>K | No | gnomAD | |
| rs1951571369 | 863 | N>H | No | Ensembl | |
| rs1452806491 | 863 | N>K | No | gnomAD | |
| rs1254976775 | 864 | E>K | No | gnomAD | |
| rs753577567 | 865 | K>E | No |
ExAC gnomAD |
|
|
COSM3908533 COSM3908532 |
866 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777107884 | 866 | R>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767684781 | 868 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1330113964 | 869 | L>R | No | gnomAD | |
| rs2132195336 | 870 | D>V | No | Ensembl | |
| rs1951569474 | 870 | D>Y | No | Ensembl | |
| rs1262040309 | 873 | I>V | No | TOPMed | |
| rs774392358 | 875 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs759775007 | 875 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1174322850 | 877 | Q>P | No | gnomAD | |
| rs148346061 | 878 | Q>E | No |
ESP gnomAD |
|
| rs937311433 | 878 | Q>R | No |
1000Genomes TOPMed gnomAD |
|
| rs1951567366 | 880 | I>L | No | TOPMed | |
| rs1411490157 | 881 | K>E | No | gnomAD | |
| rs1477895817 | 881 | K>R | No | Ensembl | |
| rs1372457518 | 882 | E>K | No | Ensembl | |
| rs554584854 | 883 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1294594800 | 884 | Q>E | No | TOPMed | |
| rs1294594800 | 884 | Q>K | No | TOPMed | |
| rs769187432 | 887 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs769187432 | 887 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs1445121618 | 888 | G>E | No |
TOPMed gnomAD |
|
| rs1588100908 | 888 | G>R | No |
TOPMed gnomAD |
|
| rs1445121618 | 888 | G>V | No |
TOPMed gnomAD |
|
| rs1186057078 | 889 | Q>E | No | gnomAD | |
| rs775221357 | 889 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs771625385 | 890 | E>K | No |
ExAC gnomAD |
|
| rs745341493 | 892 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs745341493 | 892 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1950696821 | 892 | G>V | No | Ensembl | |
|
COSM4696114 rs777539862 |
895 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs777539862 | 895 | P>R | No |
ExAC TOPMed gnomAD |
|
| COSM3996652 | 896 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752504536 | 896 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1483659279 | 897 | A>P | No | gnomAD | |
| rs1483659279 | 897 | A>T | No | gnomAD | |
| rs758582460 | 899 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1950694891 | 899 | D>Y | No | Ensembl | |
| rs750470392 | 900 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1326534482 | 902 | A>G | No |
TOPMed gnomAD |
|
| rs1347931941 | 903 | C>R | No |
TOPMed gnomAD |
|
| rs1588100396 | 906 | K>E | No | Ensembl | |
| rs1564342469 | 907 | R>M | No | Ensembl | |
| rs754449375 | 908 | R>K | No |
ExAC TOPMed gnomAD |
|
| COSM1314977 | 910 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1369860696 | 910 | G>C | No |
TOPMed gnomAD |
|
| rs1166245954 | 910 | G>V | No |
TOPMed gnomAD |
|
| TCGA novel | 911 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1950690661 | 913 | G>V | No | TOPMed | |
| rs1313346740 | 915 | I>T | No | Ensembl | |
| rs374535082 | 915 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1315855042 | 916 | D>A | No | gnomAD | |
| rs1950689528 | 917 | H>Y | No | TOPMed | |
| rs1304226749 | 918 | L>I | No |
TOPMed gnomAD |
|
| rs1304226749 | 918 | L>V | No |
TOPMed gnomAD |
|
| rs1476815611 | 919 | Q>* | No | gnomAD | |
| rs1476815611 | 919 | Q>E | No | gnomAD | |
| rs1255977149 | 919 | Q>H | No |
TOPMed gnomAD |
|
| rs767822811 | 921 | L>S | No |
ExAC gnomAD |
|
| rs760664750 | 922 | D>H | No | Ensembl | |
| TCGA novel | 923 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs3199679 | 923 | E>K | No |
ExAC gnomAD |
|
| rs1481206400 | 924 | Q>E | No | gnomAD | |
| rs2132007869 | 924 | Q>R | No | Ensembl | |
| rs1250506637 | 925 | K>E | No | Ensembl | |
|
COSM1110617 rs141746868 |
925 | K>N | endometrium [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs1058995 | 927 | W>* | No |
ExAC TOPMed gnomAD |
|
| rs770574285 | 927 | W>R | No |
ExAC gnomAD |
|
| rs772729594 | 928 | L>S | No |
ExAC gnomAD |
|
| rs769677258 | 932 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs894473317 | 932 | V>I | No | TOPMed | |
| rs1950494100 | 933 | E>Q | No | gnomAD | |
| rs1950493875 | 935 | V>I | No | Ensembl | |
| COSM3848975 | 937 | N>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM197498 rs3199677 |
939 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine central_nervous_system breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM3908531 rs566128018 |
939 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1301815809 | 940 | Q>R | No | gnomAD | |
| TCGA novel | 941 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772944172 | 941 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1397636455 | 941 | E>V | No | Ensembl | |
| rs1950492029 | 943 | E>Q | No | gnomAD | |
| rs376419864 | 944 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs376419864 | 944 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs778041126 | 944 | E>V | No |
ExAC gnomAD |
|
| rs1410215160 | 945 | L>P | No | gnomAD | |
| rs1456339726 | 945 | L>V | No | gnomAD | |
| rs1588087625 | 946 | E>D | No | Ensembl | |
| TCGA novel | 948 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1950490088 | 948 | D>N | No | TOPMed | |
| rs753202865 | 952 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs150370161 | 952 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1243771524 | 954 | A>D | No | gnomAD | |
| rs1950488288 | 955 | I>M | No | Ensembl | |
| TCGA novel | 955 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767919306 | 956 | V>F | No |
ExAC gnomAD |
|
| rs755230514 | 957 | S>F | No |
ExAC gnomAD |
|
| rs547806235 | 958 | K>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs898974916 | 960 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1317519116 | 961 | A>S | No | gnomAD | |
| rs199552193 | 961 | A>V | No |
TOPMed gnomAD |
|
| rs765966415 | 964 | Q>* | No |
ExAC gnomAD |
|
| rs762623494 | 966 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs1950485698 | 967 | S>N | No | Ensembl | |
| rs772676294 | 968 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs772676294 | 968 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs529234359 | 970 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1950484783 | 970 | E>G | No | Ensembl | |
| rs370947696 | 973 | K>T | No |
ESP TOPMed gnomAD |
|
| rs1949619359 | 979 | A>V | No | Ensembl | |
| rs796393799 | 982 | T>K | No | Ensembl | |
| rs1949617336 | 987 | I>T | No | TOPMed | |
| rs79324787 | 987 | I>V | No |
1000Genomes ESP ExAC gnomAD |
|
| rs1252514656 | 988 | S>P | No |
TOPMed gnomAD |
|
| rs748127848 | 990 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs748127848 | 990 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs146715557 | 990 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146715557 | 990 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1281839497 | 992 | N>D | No | gnomAD | |
| rs201841310 | 994 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1426447593 | 994 | L>P | No |
TOPMed gnomAD |
|
| rs201841310 | 994 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs749998476 COSM3943100 |
995 | E>Q | oesophagus [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1032402004 | 996 | Q>* | No | Ensembl | |
| rs143276779 | 996 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1391407824 | 997 | E>A | No | TOPMed | |
| rs139839095 | 997 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs117193939 | 998 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1316146865 | 1000 | E>V | No | TOPMed | |
| COSM3780147 | 1001 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760518358 | 1002 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs760518358 | 1002 | N>H | No |
ExAC TOPMed gnomAD |
|
| rs142530672 | 1002 | N>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1949612358 | 1003 | V>A | No | Ensembl | |
| rs377534036 | 1003 | V>M | No |
ESP TOPMed |
|
| rs1949612116 | 1005 | L>F | No | Ensembl | |
| rs767566638 | 1008 | S>G | No |
ExAC gnomAD |
|
| rs759346153 | 1009 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1949610997 | 1009 | T>R | No | TOPMed | |
| rs774309586 | 1010 | A>D | No |
ExAC TOPMed gnomAD |
|
| COSM3848974 | 1010 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774309586 | 1010 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs769866788 | 1012 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1437410140 | 1014 | T>R | No | TOPMed | |
|
TCGA novel rs1435939094 |
1015 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
gnomAD NCI-TCGA |
| rs1949608472 | 1016 | I>T | No | TOPMed | |
| COSM3927127 | 1017 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2131855604 | 1018 | E>K | No | Ensembl | |
| TCGA novel | 1019 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1949607615 | 1020 | V>A | No | Ensembl | |
| rs1949607376 | 1021 | E>D | No | TOPMed | |
| rs776548762 | 1021 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs776548762 | 1021 | E>Q | No |
ExAC TOPMed gnomAD |
|
| COSM487613 | 1022 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs796778171 | 1023 | L>I | No | Ensembl | |
| rs1343941050 | 1023 | L>P | No | gnomAD | |
| rs1225790587 | 1026 | E>A | No | gnomAD | |
| rs1260831650 | 1026 | E>K | No |
TOPMed gnomAD |
|
| rs1324746052 | 1027 | K>E | No |
TOPMed gnomAD |
|
| rs111906484 | 1027 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs747506325 | 1028 | D>E | No |
ExAC gnomAD |
|
| rs1265813464 | 1028 | D>N | No |
TOPMed gnomAD |
|
| rs1564316054 | 1030 | L>F | No | Ensembl | |
| rs765041527 | 1033 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs144855861 | 1033 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs144855861 | 1033 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs746194165 | 1035 | H>Q | No |
ExAC TOPMed gnomAD |
|
|
rs55654273 COSM3764032 RCV000454897 CA5102699 VAR_061286 |
1036 | N>D | large_intestine [Cosmic] | No |
ClinGen cosmic curated ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1178877821 | 1036 | N>I | No |
TOPMed gnomAD |
|
| rs1178877821 | 1036 | N>S | No |
TOPMed gnomAD |
|
| rs1429741091 | 1037 | V>L | No | gnomAD | |
| rs753586591 | 1038 | D>G | No |
ExAC gnomAD |
|
| rs756839697 | 1038 | D>N | No | ExAC | |
| rs1450841205 | 1039 | E>D | No | TOPMed | |
| COSM3375299 | 1040 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1949601520 | 1040 | K>Q | No | gnomAD | |
| rs1314967201 | 1041 | L>F | No |
TOPMed gnomAD |
|
| rs755585990 | 1042 | K>E | No |
ExAC gnomAD |
|
| rs147288435 | 1043 | N>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1949600065 | 1044 | G>D | No |
TOPMed gnomAD |
|
| TCGA novel | 1044 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1949600065 | 1044 | G>V | No |
TOPMed gnomAD |
|
| rs767513341 | 1045 | R>G | No |
ExAC gnomAD |
|
| rs201289514 | 1045 | R>T | No |
TOPMed gnomAD |
|
| rs759576962 | 1048 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs2072444257 | 1048 | S>P | No | Ensembl | |
| rs761925902 | 1049 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs761925902 | 1049 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs766244397 | 1049 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs766244397 | 1049 | P>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1051 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143915976 | 1053 | H>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143915976 | 1053 | H>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1948878824 | 1053 | H>Y | No | Ensembl | |
| rs111290584 | 1054 | V>I | No | gnomAD | |
|
COSM1212449 rs1184436591 |
1055 | L>I | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs1476150820 | 1056 | F>L | No | gnomAD | |
| rs1948877426 | 1057 | Q>* | No | Ensembl | |
| rs766080244 | 1057 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs758141623 | 1058 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1409748478 | 1059 | E>G | No |
TOPMed gnomAD |
|
| rs370735815 | 1059 | E>K | No |
TOPMed gnomAD |
|
| rs370735815 | 1059 | E>Q | No |
TOPMed gnomAD |
|
| rs2131745224 | 1060 | E>G | No | Ensembl | |
| rs369251410 | 1061 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs295274 | 1062 | I>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1948874571 | 1064 | A>T | No | Ensembl | |
| rs1948874034 | 1066 | E>G | No | gnomAD | |
| rs1455909631 | 1067 | A>T | No | TOPMed | |
| rs1337254877 | 1068 | A>E | No |
TOPMed gnomAD |
|
| rs775528711 | 1068 | A>P | No |
ExAC gnomAD |
|
| rs1337254877 | 1068 | A>V | No |
TOPMed gnomAD |
|
| rs371974528 | 1069 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs767645576 | 1069 | I>V | No |
ExAC gnomAD |
|
| TCGA novel | 1070 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948871669 | 1072 | R>K | No | TOPMed | |
| rs774856531 | 1073 | N>K | No |
ExAC gnomAD |
|
| rs1948871132 | 1074 | E>G | No | Ensembl | |
| rs749616984 | 1076 | I>N | No |
ExAC gnomAD |
|
| rs1404071284 | 1078 | N>Y | No | gnomAD | |
| rs773480141 | 1079 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs950185206 | 1079 | R>H | No |
TOPMed gnomAD |
|
| rs950185206 | 1079 | R>L | No |
TOPMed gnomAD |
|
| TCGA novel | 1080 | Q>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2131744161 | 1082 | S>* | No | Ensembl | |
| rs879094423 | 1083 | L>F | No | Ensembl | |
| rs1425175132 | 1083 | L>H | No | gnomAD | |
| rs1350191666 | 1084 | R>T | No |
TOPMed gnomAD |
|
| rs1353417932 | 1085 | A>T | No | gnomAD | |
| rs1948868448 | 1085 | A>V | No | Ensembl | |
| rs917532006 | 1088 | H>R | No | TOPMed | |
| rs1411363040 | 1088 | H>Y | No | gnomAD | |
| rs1948867169 | 1089 | N>I | No | TOPMed | |
| TCGA novel | 1091 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs529481603 COSM1110616 |
1092 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs529481603 | 1092 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs747850098 | 1092 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs747850098 | 1092 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs529481603 | 1092 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1948865876 | 1093 | G>S | No | Ensembl | |
| rs1948865067 | 1094 | E>* | No |
TOPMed gnomAD |
|
| rs1948865067 | 1094 | E>K | No |
TOPMed gnomAD |
|
| rs780834995 | 1095 | A>E | No | ExAC | |
| TCGA novel | 1095 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs565946178 | 1096 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs779980486 | 1100 | K>E | No |
ExAC gnomAD |
|
| rs1206706411 | 1101 | L>I | No |
TOPMed gnomAD |
|
| rs1206706411 | 1101 | L>V | No |
TOPMed gnomAD |
|
| rs1308376362 | 1102 | A>V | No | gnomAD | |
| rs1948861881 | 1103 | C>F | No | gnomAD | |
| rs756317575 | 1105 | S>C | No |
ExAC gnomAD |
|
| rs752987077 | 1105 | S>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1107 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1203198561 | 1109 | I>M | No | TOPMed | |
| rs1443702696 | 1110 | R>G | No |
TOPMed gnomAD |
|
| rs1948859437 | 1110 | R>S | No | Ensembl | |
| rs1354803310 | 1110 | R>T | No |
TOPMed gnomAD |
|
| rs909861058 | 1111 | T>A | No | TOPMed | |
| rs759556164 | 1112 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs759556164 | 1112 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1948858263 | 1113 | L>I | No |
TOPMed gnomAD |
|
| rs531876952 | 1114 | F>L | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 1118 | N>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763409241 | 1118 | N>S | No |
ExAC gnomAD |
|
| rs1948395100 | 1120 | V>M | No | Ensembl | |
| rs772028568 | 1121 | V>L | No |
ExAC gnomAD |
|
| rs1237447458 | 1123 | L>V | No |
TOPMed gnomAD |
|
| rs1303828659 | 1124 | R>* | No |
TOPMed gnomAD |
|
| rs1303828659 | 1124 | R>G | No |
TOPMed gnomAD |
|
| rs745853642 | 1124 | R>Q | No |
ExAC gnomAD |
|
| rs778677386 | 1125 | E>Q | No |
ExAC gnomAD |
|
| rs1390164598 | 1126 | A>G | No | gnomAD | |
| rs1420165059 | 1127 | E>K | No | gnomAD | |
| rs368566567 | 1128 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs756981018 | 1128 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1948391802 | 1129 | K>T | No | Ensembl | |
| rs778425748 | 1130 | Q>R | No | Ensembl | |
| rs1446716258 | 1131 | Q>* | No | gnomAD | |
| rs1948389914 | 1132 | L>* | No | Ensembl | |
| rs1200882258 | 1132 | L>V | No | gnomAD | |
| rs141429420 | 1133 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1055187908 | 1133 | Y>C | No |
TOPMed gnomAD |
|
| rs781343039 | 1133 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1354264977 | 1134 | N>S | No | gnomAD | |
| COSM1212448 | 1136 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1136 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1243092484 | 1136 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 1138 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1384622492 | 1139 | M>I | No |
TOPMed gnomAD |
|
| TCGA novel | 1139 | M>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948387283 | 1142 | L>R | No |
TOPMed gnomAD |
|
| rs763502349 | 1144 | R>L | No |
ExAC gnomAD |
|
| rs763502349 | 1144 | R>Q | No |
ExAC gnomAD |
|
|
COSM456163 rs370478477 |
1144 | R>W | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1383304001 | 1145 | D>N | No | gnomAD | |
| rs751037465 | 1146 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1948385156 | 1147 | M>I | No | TOPMed | |
|
rs58077086 CA5102618 RCV000455471 |
1147 | M>V | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1948384892 | 1148 | V>I | No | TOPMed | |
| rs762208623 | 1149 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs762208623 | 1149 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs377569727 | 1149 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs377569727 | 1149 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1444177147 | 1151 | L>* | No |
TOPMed gnomAD |
|
| rs775056643 | 1152 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs375552082 | 1153 | S>P | No |
ESP ExAC gnomAD |
|
| rs1426920114 | 1154 | A>V | No | gnomAD | |
| rs1259424425 | 1155 | L>P | No | gnomAD | |
| rs1384078903 | 1155 | L>V | No |
TOPMed gnomAD |
|
| rs1450639451 | 1156 | D>N | No | TOPMed | |
|
rs1948380493 TCGA novel |
1161 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs749043182 | 1162 | C>G | No |
ExAC gnomAD |
|
| rs749043182 | 1162 | C>R | No |
ExAC gnomAD |
|
| rs71502726 | 1164 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs71502726 | 1164 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs377744428 | 1164 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780176063 | 1168 | L>H | No |
ExAC gnomAD |
|
| rs1948378136 | 1169 | Q>R | No |
TOPMed gnomAD |
|
| rs201858465 | 1170 | Q>L | No |
TOPMed gnomAD |
|
| rs201858465 | 1170 | Q>R | No |
TOPMed gnomAD |
|
| rs750542303 | 1172 | E>K | No |
ExAC TOPMed gnomAD |
|
|
rs1391582977 COSM3930166 |
1174 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1391582977 | 1174 | E>Q | No |
TOPMed gnomAD |
|
| rs955291544 | 1175 | Q>* | No |
TOPMed gnomAD |
|
| rs955291544 | 1175 | Q>E | No |
TOPMed gnomAD |
|
| rs955291544 | 1175 | Q>K | No |
TOPMed gnomAD |
|
| rs371296942 | 1178 | Q>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1406375197 | 1178 | Q>H | No | TOPMed | |
| rs764428820 | 1178 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 1180 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1022499611 | 1181 | L>* | No | TOPMed | |
| rs1948373003 | 1182 | H>Y | No | TOPMed | |
| TCGA novel | 1183 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759000017 | 1183 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1267501956 | 1184 | F>L | No | gnomAD | |
| rs544615598 | 1184 | F>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1701265 rs1948371357 |
1186 | E>K | skin [Cosmic] | No |
cosmic curated TOPMed |
| rs1400392636 | 1191 | G>C | No | TOPMed | |
| rs765929828 | 1192 | I>S | No |
ExAC gnomAD |
|
| rs762873465 | 1195 | T>S | No |
ExAC gnomAD |
|
| rs1365751041 | 1196 | F>L | No |
TOPMed gnomAD |
|
| rs1298457265 | 1196 | F>Y | No | Ensembl | |
| rs554621908 | 1198 | T>K | No |
1000Genomes ExAC |
|
| rs1414500674 | 1199 | Y>* | No | gnomAD | |
| rs1946673147 | 1199 | Y>C | No | TOPMed | |
| rs747845313 | 1199 | Y>H | No |
ExAC gnomAD |
|
| rs776518666 | 1200 | E>A | No |
ExAC gnomAD |
|
| rs772150791 | 1201 | D>N | No | ExAC | |
| rs779132941 | 1202 | K>N | No |
ExAC gnomAD |
|
| rs749396761 | 1205 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs757343170 | 1205 | Q>P | No |
ExAC gnomAD |
|
| rs1946670905 | 1207 | E>Q | No |
TOPMed gnomAD |
|
| rs778094907 | 1207 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1402572716 | 1209 | D>H | No | gnomAD | |
| rs1402572716 | 1209 | D>N | No | gnomAD | |
| rs1946670062 | 1210 | L>H | No | TOPMed | |
| rs1331838704 | 1211 | Y>* | No |
TOPMed gnomAD |
|
| rs756676470 | 1211 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs752975744 | 1213 | Y>C | No |
ExAC gnomAD |
|
| rs1187220292 | 1213 | Y>H | No |
TOPMed gnomAD |
|
| COSM422329 | 1214 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3848973 | 1214 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs542484683 | 1216 | T>I | No |
1000Genomes ExAC gnomAD |
|
| rs1434097150 | 1218 | R>Q | No | TOPMed | |
|
COSM3908530 rs376993923 |
1218 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1178714065 | 1220 | H>D | No |
TOPMed gnomAD |
|
| rs1212749655 | 1221 | K>* | No | gnomAD | |
| rs1946666295 | 1221 | K>T | No | Ensembl | |
| COSM1110615 | 1222 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1946665841 | 1222 | K>R | No | Ensembl | |
| rs200646527 | 1223 | K>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 1223 | K>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762391417 | 1228 | V>A | No |
ExAC gnomAD |
|
| rs1390923807 | 1228 | V>I | No |
TOPMed gnomAD |
|
| rs1946663476 | 1230 | E>* | No | TOPMed | |
| rs146464985 | 1233 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1317446725 COSM261665 |
1233 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs761847847 | 1234 | R>L | No |
ExAC TOPMed gnomAD |
|
|
rs761847847 COSM4928715 |
1234 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs368997037 | 1234 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1054489880 | 1238 | P>S | No | gnomAD | |
| rs199566689 | 1241 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1166269708 | 1242 | Q>H | No |
TOPMed gnomAD |
|
| rs2542780 | 1243 | E>K | No |
TOPMed gnomAD |
|
| rs1418348597 | 1246 | D>G | No |
TOPMed gnomAD |
|
| rs919485448 | 1247 | G>R | No | TOPMed | |
| rs1946010451 | 1250 | K>Q | No | TOPMed | |
| rs1350413295 | 1252 | E>G | No |
TOPMed gnomAD |
|
| COSM4897076 | 1253 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1564256360 | 1256 | M>L | No | Ensembl | |
| rs1946005577 | 1258 | S>L | No | Ensembl | |
| rs1489988620 | 1259 | E>K | No | gnomAD | |
| rs1248579450 | 1259 | E>V | No | gnomAD | |
| rs149899595 | 1260 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs753745000 | 1261 | L>* | No |
ExAC gnomAD |
|
| rs764144016 | 1262 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs1946002046 | 1263 | W>* | No | Ensembl | |
| rs1946001522 | 1265 | S>C | No | TOPMed | |
| rs1946000518 | 1267 | P>A | No | Ensembl | |
| rs775457038 | 1268 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs540787693 | 1268 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs540787693 | 1268 | E>Q | No |
1000Genomes ExAC gnomAD |
|
| rs771248074 | 1269 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs771248074 | 1269 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1278556818 | 1270 | M>V | No |
TOPMed gnomAD |
|
| rs2131425507 | 1271 | K>T | No | Ensembl | |
| TCGA novel | 1272 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1276 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs528796605 | 1276 | E>G | No |
1000Genomes ExAC gnomAD |
|
| rs2131425411 | 1277 | R>T | No | Ensembl | |
| rs201708301 | 1279 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1945995793 | 1282 | S>A | No | Ensembl | |
| rs1294847517 | 1284 | S>G | No | gnomAD | |
| rs1945994223 | 1285 | S>G | No | Ensembl | |
| rs1945993159 | 1286 | L>F | No |
TOPMed gnomAD |
|
| rs769982330 | 1286 | L>V | No |
ExAC gnomAD |
|
| rs1458513399 | 1287 | R>K | No | TOPMed | |
| rs748179128 | 1288 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1362287325 | 1289 | Q>R | No |
TOPMed gnomAD |
|
|
rs781620659 COSM4164069 |
1292 | P>L | thyroid [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1466826673 | 1293 | Q>* | No |
TOPMed gnomAD |
|
| rs1945991069 | 1293 | Q>R | No | Ensembl | |
| rs1469642275 | 1296 | W>R | No | TOPMed | |
| rs769173452 | 1297 | E>K | No |
ExAC gnomAD |
|
| rs1176133064 | 1298 | D>A | No | gnomAD | |
| rs140117578 | 1299 | I>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs140117578 | 1299 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1945986781 | 1300 | P>Q | No | Ensembl | |
| rs561598508 | 1300 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs561598508 | 1300 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1301 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2131424223 | 1303 | P>L | No | Ensembl | |
| rs1393403730 | 1305 | I>V | No | TOPMed | |
| rs1945985728 | 1306 | H>Y | No | Ensembl | |
| TCGA novel | 1307 | S>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1945985190 | 1307 | S>G | No | Ensembl | |
| rs778438479 | 1307 | S>I | No |
ExAC gnomAD |
|
| rs778438479 | 1307 | S>T | No |
ExAC gnomAD |
|
| rs756736442 | 1308 | S>F | No |
ExAC TOPMed gnomAD |
|
| COSM261664 | 1308 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945983442 | 1309 | L>S | No | Ensembl | |
| rs764086639 | 1311 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs764086639 | 1311 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs764086639 | 1311 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs756162275 | 1312 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs756162275 | 1312 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs756162275 | 1312 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1458343983 | 1312 | P>S | No | gnomAD | |
| TCGA novel | 1313 | S>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs775622981 COSM6039561 |
1313 | S>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs752519365 | 1314 | G>E | No |
ExAC gnomAD |
|
| rs767480512 | 1315 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1372024316 | 1316 | M>I | No |
TOPMed gnomAD |
|
| rs1159901509 | 1318 | G>R | No | gnomAD | |
| rs1159901509 | 1318 | G>S | No | gnomAD | |
| rs763083436 | 1319 | N>Y | No |
ExAC TOPMed gnomAD |
|
| rs765389677 | 1320 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs761903243 | 1321 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs1945973399 | 1324 | E>Q | No | gnomAD | |
| rs769116359 | 1325 | T>I | No |
ExAC gnomAD |
|
| rs769116359 | 1325 | T>R | No |
ExAC gnomAD |
|
| rs1437459098 | 1327 | D>G | No | Ensembl | |
| rs1361201084 | 1327 | D>Y | No | gnomAD | |
| rs1945968904 | 1328 | N>H | No | gnomAD | |
| rs1418960058 | 1329 | Q>H | No | gnomAD | |
| rs747561292 | 1331 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs775815260 | 1332 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1482156736 | 1333 | S>C | No | gnomAD | |
| rs1482156736 | 1333 | S>F | No | gnomAD | |
| rs541489156 | 1334 | H>Q | No |
1000Genomes ExAC gnomAD |
|
| rs772417015 | 1334 | H>Y | No |
ExAC gnomAD |
|
| rs371473677 | 1336 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs369880739 | 1336 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs746336857 | 1338 | S>T | No | Ensembl | |
| rs1390436829 | 1339 | S>C | No |
TOPMed gnomAD |
|
| rs1390436829 | 1339 | S>F | No |
TOPMed gnomAD |
|
| rs1319020945 | 1340 | Q>* | No |
TOPMed gnomAD |
|
| rs1319020945 | 1340 | Q>E | No |
TOPMed gnomAD |
|
| rs1377255524 | 1341 | I>T | No | gnomAD | |
| rs1356773992 | 1342 | Q>* | No |
TOPMed gnomAD |
|
| rs1412392606 | 1342 | Q>H | No | TOPMed | |
| rs1945957026 | 1342 | Q>P | No | Ensembl | |
| rs1319199604 | 1343 | V>A | No | TOPMed | |
| rs748776654 | 1343 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs1393862735 | 1348 | G>E | No | gnomAD | |
| rs1221074580 | 1349 | R>* | No | Ensembl | |
| rs777475564 | 1349 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1249944935 | 1350 | L>V | No |
TOPMed gnomAD |
|
| rs904928731 | 1351 | H>L | No | TOPMed | |
| rs767566257 | 1352 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs767566257 | 1352 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1945950524 | 1353 | V>A | No | gnomAD | |
| rs1484699358 | 1355 | P>R | No | TOPMed | |
| rs574356626 | 1355 | P>S | No |
1000Genomes TOPMed gnomAD |
|
| rs1945948732 | 1359 | C>R | No | Ensembl | |
| rs1420401209 | 1359 | C>Y | No | gnomAD | |
|
COSM3220258 rs375822223 |
1360 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs761977367 | 1360 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs1425730729 COSM3433307 |
1362 | E>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| rs1425730729 | 1362 | E>Q | No |
TOPMed gnomAD |
|
| rs1258071081 | 1364 | R>C | No |
TOPMed gnomAD |
|
| rs1292479233 | 1364 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM3659200 | 1367 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs376463665 | 1368 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs376463665 | 1368 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1945942012 | 1369 | L>S | No | Ensembl | |
| rs1945942383 | 1369 | L>V | No | TOPMed | |
| rs746207649 | 1370 | E>K | No | ExAC | |
| rs770595664 | 1371 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1242183798 | 1372 | S>* | No |
TOPMed gnomAD |
|
|
rs369241220 COSM3726555 |
1374 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1945939519 | 1374 | R>L | No | Ensembl | |
| rs1381997894 | 1375 | R>C | No | gnomAD | |
| rs777419059 | 1375 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1587833398 | 1377 | S>G | No | gnomAD | |
| rs1587833398 | 1377 | S>R | No | gnomAD | |
| rs1945935282 | 1380 | V>A | No | gnomAD | |
| rs142462766 | 1380 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1432436456 | 1384 | S>A | No | gnomAD | |
| rs932090047 | 1385 | M>V | No | TOPMed | |
| rs747672794 | 1386 | A>T | No |
ExAC gnomAD |
|
| rs1692312370 | 1388 | D>E | No | TOPMed | |
| rs1459679335 | 1389 | S>C | No | gnomAD | |
| COSM3927126 | 1389 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1161515061 | 1389 | S>P | No | gnomAD | |
| rs375179706 | 1391 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs140974886 | 1393 | S>Y | No |
ESP TOPMed gnomAD |
|
| rs1945930557 | 1396 | P>L | No | TOPMed | |
| rs1945931268 | 1396 | P>T | No | Ensembl | |
| rs1195793051 | 1397 | R>K | No | gnomAD | |
| rs1195793051 | 1397 | R>T | No | gnomAD | |
| COSM1110614 | 1398 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945929217 | 1398 | D>Y | No | Ensembl | |
| TCGA novel | 1400 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1267613573 | 1400 | K>T | No |
TOPMed gnomAD |
|
| rs779758665 | 1401 | T>I | No |
ExAC gnomAD |
No associated diseases with Q86VH2
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cilium | A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface and of some cytoplasmic parts. Each cilium is largely bounded by an extrusion of the cytoplasmic (plasma) membrane, and contains a regular longitudinal array of microtubules, anchored to a basal body. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| kinesin complex | Any complex that includes a dimer of molecules from the kinesin superfamily, a group of related proteins that contain an extended region of predicted alpha-helical coiled coil in the main chain that likely produces dimerization. The native complexes of several kinesin family members have also been shown to contain additional peptides, often designated light chains as all of the noncatalytic subunits that are currently known are smaller than the chain that contains the motor unit. Kinesin complexes generally possess a force-generating enzymatic activity, or motor, which converts the free energy of the gamma phosphate bond of ATP into mechanical work. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| microtubule motor activity | A motor activity that generates movement along a microtubule, driven by ATP hydrolysis. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cilium assembly | The assembly of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole. |
| epithelial cilium movement involved in extracellular fluid movement | The directed, self-propelled movement of cilia of epithelial cells. Depending on the type of cell, there may be one or many cilia per cell. This movement is usually coordinated between many epithelial cells, and serves to move extracellular fluid. |
| establishment of localization in cell | Any process, occuring in a cell, that localizes a substance or cellular component. This may occur via movement, tethering or selective degradation. |
| microtubule-based movement | A microtubule-based process that results in the movement of organelles, other microtubules, or other cellular components. Examples include motor-driven movement along microtubules and movement driven by polymerization or depolymerization of microtubules. |
| mitotic spindle organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the microtubule spindle during a mitotic cell cycle. |
| spindle elongation | The cell cycle process in which the distance is lengthened between poles of the spindle. |
| ventricular system development | The process whose specific outcome is the progression of the brain ventricular system over time, from its formation to the mature structure. The brain ventricular system consists of four communicating cavities within the brain that are continuous with the central canal of the spinal cord. These cavities include two lateral ventricles, the third ventricle and the fourth ventricle. Cerebrospinal fluid fills the ventricles and is produced by the choroid plexus. |
20 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O95239 | KIF4A | Chromosome-associated kinesin KIF4A | Homo sapiens (Human) | SS |
| Q2VIQ3 | KIF4B | Chromosome-associated kinesin KIF4B | Homo sapiens (Human) | SS |
| P33176 | KIF5B | Kinesin-1 heavy chain | Homo sapiens (Human) | EV |
| O60282 | KIF5C | Kinesin heavy chain isoform 5C | Homo sapiens (Human) | EV |
| Q12840 | KIF5A | Kinesin heavy chain isoform 5A | Homo sapiens (Human) | EV |
| Q2M1P5 | KIF7 | Kinesin-like protein KIF7 | Homo sapiens (Human) | EV |
| Q7Z4S6 | KIF21A | Kinesin-like protein KIF21A | Homo sapiens (Human) | EV |
| O75037 | KIF21B | Kinesin-like protein KIF21B | Homo sapiens (Human) | EV |
| Q9P2E2 | KIF17 | Kinesin-like protein KIF17 | Homo sapiens (Human) | EV |
| O00139 | KIF2A | Kinesin-like protein KIF2A | Homo sapiens (Human) | PR |
| Q9NQT8 | KIF13B | Kinesin-like protein KIF13B | Homo sapiens (Human) | EV |
| Q9H1H9 | KIF13A | Kinesin-like protein KIF13A | Homo sapiens (Human) | SS |
| O43896 | KIF1C | Kinesin-like protein KIF1C | Homo sapiens (Human) | SS |
| Q9QXL1 | Kif21b | Kinesin-like protein KIF21B | Mus musculus (Mouse) | SS |
| Q9QXL2 | Kif21a | Kinesin-like protein KIF21A | Mus musculus (Mouse) | EV SS |
| B7ZNG0 | Kif7 | Kinesin-like protein KIF7 | Mus musculus (Mouse) | SS |
| Q7M6Z4 | Kif27 | Kinesin-like protein KIF27 | Mus musculus (Mouse) | SS |
| F1M5N7 | Kif21b | Kinesin-like protein KIF21B | Rattus norvegicus (Rat) | SS |
| Q7M6Z5 | Kif27 | Kinesin-like protein KIF27 | Rattus norvegicus (Rat) | SS |
| Q58G59 | kif7 | Kinesin-like protein kif7 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEEIPVKVAV | RIRPLLCKEA | LHNHQVCVRV | IPNSQQVIIG | RDRVFTFDFV | FGKNSTQDEV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YNTCIKPLVL | SLIEGYNATV | FAYGQTGSGK | TYTIGGGHIA | SVVEGQKGII | PRAIQEIFQS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ISEHPSIDFN | VKVSYIEVYK | EDLRDLLELE | TSMKDLHIRE | DEKGNTVIVG | AKECHVESAG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EVMSLLEMGN | AARHTGTTQM | NEHSSRSHAI | FTISICQVHK | NMEAAEDGSW | YSPRHIVSKF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| HFVDLAGSER | VTKTGNTGER | FKESIQINSG | LLALGNVISA | LGDPRRKSSH | IPYRDAKITR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LLKDSLGGSA | KTVMITCVSP | SSSNFDESLN | SLKYANRARN | IRNKPTVNFS | PESDRIDEME |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FEIKLLREAL | QSQQAGVSQT | TQINREGSPD | TNRIHSLEEQ | VAQLQGECLG | YQCCVEEAFT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FLVDLKDTVR | LNEKQQHKLQ | EWFNMIQEVR | KAVLTSFRGI | GGTASLEEGP | QHVTVLQLKR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ELKKCQCVLA | ADEVVFNQKE | LEVKELKNQV | QMMVQENKGH | AVSLKEAQKV | NRLQNEKIIE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| QQLLVDQLSE | ELTKLNLSVT | SSAKENCGDG | PDARIPERRP | YTVPFDTHLG | HYIYIPSRQD |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SRKVHTSPPM | YSLDRIFAGF | RTRSQMLLGH | IEEQDKVLHC | QFSDNSDDEE | SEGQEKSGTR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| CRSRSWIQKP | DSVCSLVELS | DTQDETQKSD | LENEDLKIDC | LQESQELNLQ | KLKNSERILT |
| 730 | 740 | 750 | 760 | 770 | 780 |
| EAKQKMRELT | INIKMKEDLI | KELIKTGNDA | KSVSKQYSLK | VTKLEHDAEQ | AKVELIETQK |
| 790 | 800 | 810 | 820 | 830 | 840 |
| QLQELENKDL | SDVAMKVKLQ | KEFRKKMDAA | KLRVQVLQKK | QQDSKKLASL | SIQNEKRANE |
| 850 | 860 | 870 | 880 | 890 | 900 |
| LEQSVDHMKY | QKIQLQRKLR | EENEKRKQLD | AVIKRDQQKI | KEIQLKTGQE | EGLKPKAEDL |
| 910 | 920 | 930 | 940 | 950 | 960 |
| DACNLKRRKG | SFGSIDHLQK | LDEQKKWLDE | EVEKVLNQRQ | ELEELEADLK | KREAIVSKKE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| ALLQEKSHLE | NKKLRSSQAL | NTDSLKISTR | LNLLEQELSE | KNVQLQTSTA | EEKTKISEQV |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| EVLQKEKDQL | QKRRHNVDEK | LKNGRVLSPE | EEHVLFQLEE | GIEALEAAIE | YRNESIQNRQ |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| KSLRASFHNL | SRGEANVLEK | LACLSPVEIR | TILFRYFNKV | VNLREAERKQ | QLYNEEMKMK |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| VLERDNMVRE | LESALDHLKL | QCDRRLTLQQ | KEHEQKMQLL | LHHFKEQDGE | GIMETFKTYE |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| DKIQQLEKDL | YFYKKTSRDH | KKKLKELVGE | AIRRQLAPSE | YQEAGDGVLK | PEGGGMLSEE |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| LKWASRPESM | KLSGREREMD | SSASSLRTQP | NPQKLWEDIP | ELPPIHSSLA | PPSGHMLGNE |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| NKTETDDNQF | TKSHSRLSSQ | IQVVGNVGRL | HGVTPVKLCR | KELRQISALE | LSLRRSSLGV |
| 1390 | 1400 | ||||
| GIGSMAADSI | EVSRKPRDLK | T |