Q86UV7
Gene name |
TRIM73 (TRIM50B) |
Protein name |
Tripartite motif-containing protein 73 |
Names |
Tripartite motif-containing protein 50B |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:375593 |
EC number |
|
Protein Class |
|
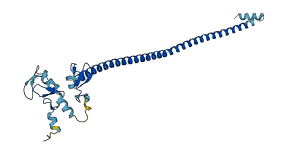
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q86UV7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q86UV7-F1 | Predicted | AlphaFoldDB |
212 variants for Q86UV7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1481786894 CA367720564 |
3 | W>C | No |
ClinGen TOPMed |
|
|
rs1554467427 CA367720545 |
3 | W>R | No |
ClinGen gnomAD |
|
|
CA4299792 rs146512104 |
4 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs371153005 CA4299793 |
5 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs73702636 CA4299795 |
8 | L>P | Variant assessed as Somatic; 0.004289 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA367720682 rs4354242 |
8 | L>V | No |
ClinGen Ensembl |
|
| TCGA novel | 9 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA367720744 rs1584621239 |
10 | L>V | No |
ClinGen Ensembl |
|
|
CA4299798 rs782489803 CA367720804 |
12 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs1554467435 CA367720783 |
12 | D>H | No |
ClinGen gnomAD |
|
|
CA4299799 rs138355430 |
13 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA159886326 rs139576761 |
13 | R>W | No |
ClinGen 1000Genomes |
|
|
CA367720851 rs1554467440 |
14 | L>P | No |
ClinGen gnomAD |
|
|
CA367720892 rs1394007021 |
16 | C>G | No |
ClinGen TOPMed |
|
|
CA367720899 rs1297591537 |
16 | C>Y | No |
ClinGen TOPMed |
|
|
rs1554467442 CA367720916 |
17 | P>A | No |
ClinGen gnomAD |
|
|
rs782548237 CA4299801 |
17 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA4299804 rs782382034 |
19 | C>F | No |
ClinGen ExAC gnomAD |
|
|
CA4299803 rs236632 |
19 | C>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA367720958 rs782382034 |
19 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs374449175 CA159886343 |
21 | E>Q | No |
ClinGen 1000Genomes gnomAD |
|
|
CA4299808 rs371634793 |
23 | F>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs782157548 CA4299810 |
24 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs782405921 CA4299811 |
24 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1554467448 CA367721150 |
25 | E>D | No |
ClinGen gnomAD |
|
|
rs782003042 CA4299812 |
25 | E>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765433508 CA4299813 |
26 | S>P | No |
ClinGen ExAC gnomAD |
|
|
rs1554467449 CA367721187 |
27 | L>Q | No |
ClinGen gnomAD |
|
|
rs781818109 CA4299815 |
28 | M>K | No |
ClinGen ExAC gnomAD |
|
|
rs781818109 CA367721213 |
28 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA367721256 rs1195768177 |
30 | Q>* | No |
ClinGen TOPMed |
|
|
CA367721254 rs1195768177 |
30 | Q>E | No |
ClinGen TOPMed |
|
|
CA4299817 rs782701009 |
30 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs143976895 CA4299820 |
31 | C>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs781889944 CA4299818 |
31 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA367721302 rs1554467455 |
32 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs781850179 CA4299821 |
32 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1554467458 CA367721351 |
34 | S>Y | No |
ClinGen gnomAD |
|
|
CA4299823 rs782588789 |
38 | G>C | No |
ClinGen ExAC gnomAD |
|
|
rs1554467469 CA367721470 |
39 | C>F | No |
ClinGen gnomAD |
|
|
CA4299825 rs782420503 |
43 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1306018926 CA367721558 |
44 | S>F | No |
ClinGen TOPMed |
|
|
CA4299826 rs782671348 |
45 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1387343931 CA367721569 |
45 | Y>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1387343931 CA367721566 |
45 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1323737040 CA367721594 |
46 | H>Y | No |
ClinGen TOPMed |
|
|
rs782255322 CA4299827 |
47 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA367721629 rs782374707 |
48 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4299828 rs782374707 |
48 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1159411951 CA367721656 |
49 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs781973423 CA4299829 |
50 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA4299830 rs782076549 |
51 | V>A | No |
ClinGen ExAC gnomAD |
|
|
COSM1091732 COSM1091733 rs1362484740 CA367721723 |
52 | R>C | Variant assessed as Somatic; 5.463e-05 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA367721721 rs1362484740 |
52 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs782323591 CA4299831 |
52 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs1362484740 CA367721718 |
52 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1554467493 CA367721783 |
54 | P>L | No |
ClinGen gnomAD |
|
|
CA367721799 rs1284312224 CA367721802 |
55 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA4299833 rs587744714 |
55 | M>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1206596661 CA367721811 |
56 | C>G | No |
ClinGen TOPMed |
|
|
CA367721842 rs1554467498 |
57 | W>* | No |
ClinGen gnomAD |
|
|
rs1348085069 CA367721835 |
57 | W>R | No |
ClinGen TOPMed |
|
|
CA367721865 rs1554467500 |
58 | Q>* | No |
ClinGen gnomAD |
|
|
rs1259726528 CA367721900 |
59 | V>A | No |
ClinGen TOPMed |
|
|
rs781860895 CA4299835 |
61 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs587646349 CA4299837 |
62 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA367721953 rs587646349 |
62 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4299838 rs782730971 |
63 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1301190912 CA367721967 |
63 | S>R | No |
ClinGen TOPMed |
|
|
rs1554467514 CA367722009 |
64 | S>N | No |
ClinGen Ensembl |
|
|
rs1554467512 CA367721994 |
64 | S>R | No |
ClinGen Ensembl |
|
|
CA367722044 rs1554467516 |
66 | L>F | No |
ClinGen gnomAD |
|
|
CA367722041 rs1584621654 |
66 | L>W | No |
ClinGen Ensembl |
|
|
rs781808072 CA4299839 |
68 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4299840 rs782437807 |
69 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM248708 rs199563025 COSM248707 CA4299841 |
73 | W>R | pancreas central_nervous_system [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs781881696 CA4299842 |
73 | W>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1454217789 CA367722195 |
75 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1584621702 CA367722217 |
76 | E>D | No |
ClinGen Ensembl |
|
|
CA367722199 rs1194717153 |
76 | E>K | No |
ClinGen TOPMed |
|
|
CA367722254 rs1584621710 |
77 | A>V | No |
ClinGen Ensembl |
|
|
CA367722270 rs1584621720 |
79 | R>G | No |
ClinGen Ensembl |
|
|
CA367722300 rs1260648400 |
80 | L>F | No |
ClinGen TOPMed |
|
|
rs1554467529 CA367722332 |
81 | P>R | No |
ClinGen gnomAD |
|
|
rs1211101438 CA367722326 |
81 | P>S | No |
ClinGen TOPMed |
|
|
rs1584621745 CA367722374 |
83 | D>A | No |
ClinGen Ensembl |
|
|
rs1584621742 CA367722370 |
83 | D>H | No |
ClinGen Ensembl |
|
|
rs1584621742 CA367722368 |
83 | D>Y | No |
ClinGen Ensembl |
|
|
rs1349843928 CA367722397 |
84 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1349843928 CA367722395 |
84 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1584621766 CA367722434 |
86 | P>L | No |
ClinGen Ensembl |
|
|
rs1233555861 CA367722426 |
86 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA367722445 rs1339974265 |
87 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA367722458 rs1271800198 |
88 | V>A | No |
ClinGen TOPMed |
|
|
CA367722492 rs1324718707 |
90 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA4299844 rs782624070 |
91 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367722524 rs1554467543 |
92 | H>P | No |
ClinGen gnomAD |
|
|
rs1415594818 CA367722536 |
93 | R>G | No |
ClinGen TOPMed |
|
|
COSM3412292 CA367722541 rs1311429469 COSM3412293 |
93 | R>Q | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
COSM3833240 CA367722537 rs1415594818 COSM3833241 |
93 | R>W | breast [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA367722554 rs1554467548 |
94 | N>K | No |
ClinGen gnomAD |
|
|
rs1420858631 CA367722566 |
95 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1474155231 CA367722569 |
96 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1474155231 CA367722568 |
96 | L>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1188261638 CA367722589 |
98 | L>F | No |
ClinGen TOPMed |
|
|
rs1486443645 CA367722598 |
99 | F>L | No |
ClinGen TOPMed |
|
|
CA367722599 rs1486443645 |
99 | F>V | No |
ClinGen TOPMed |
|
|
CA4299845 rs782226889 |
100 | C>R | No |
ClinGen ExAC gnomAD |
|
|
CA159886484 rs587619331 |
101 | E>D | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA367722669 rs1438120645 |
102 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
CA367722697 rs1554467554 |
104 | Q>R | No |
ClinGen gnomAD |
|
|
CA4299847 rs782579602 |
107 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs1554467562 CA367722789 |
109 | G>V | No |
ClinGen gnomAD |
|
|
CA367722829 rs782420159 |
111 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1337579676 CA367722839 |
112 | G>S | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 114 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1277643064 CA367722871 |
114 | L>Q | No |
ClinGen TOPMed |
|
|
CA367722937 rs1554467572 |
117 | H>R | No |
ClinGen TOPMed |
|
|
CA367722976 rs1554467576 |
118 | Q>H | No |
ClinGen gnomAD |
|
|
rs1234193509 CA367723139 |
121 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA367723136 rs1234193509 |
121 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1291599474 CA367723153 |
122 | V>I | No |
ClinGen TOPMed |
|
|
rs782004984 CA4299850 |
123 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367723211 rs1164987529 |
125 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA367723244 rs1475867112 |
126 | S>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA4299852 rs199603365 |
128 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4299853 rs367761658 |
129 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367723361 rs1202437386 |
131 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1437153352 CA367723364 |
131 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA367723368 rs1437153352 |
131 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1437153352 CA367723366 |
131 | R>P | No |
ClinGen TOPMed gnomAD |
|
|
CA367723374 rs1214404535 |
132 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1214404535 CA367723376 |
132 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1554467595 CA367723403 |
133 | K>T | No |
ClinGen gnomAD |
|
|
rs1584624858 CA367723769 |
158 | V>G | No |
ClinGen Ensembl |
|
|
CA159886575 COSM117118 rs200694955 |
159 | K>N | ovary [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs199982097 COSM287717 COSM287718 CA159886581 |
163 | R>* | kidney large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes |
|
rs1256093622 CA367723920 |
169 | D>H | No |
ClinGen TOPMed |
|
|
CA4299858 rs782766081 |
173 | W>* | No |
ClinGen ExAC |
|
|
CA367724015 rs1179785203 |
176 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1179785203 CA367724013 |
176 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA4299859 rs781848652 |
176 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367724017 rs1179785203 |
176 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs782472919 CA4299861 |
177 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782472919 CA4299860 |
177 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA159886614 rs587732630 |
177 | R>H | No |
ClinGen 1000Genomes gnomAD |
|
|
CA367724028 rs587732630 |
177 | R>L | No |
ClinGen 1000Genomes gnomAD |
|
|
rs587626362 CA4299864 |
178 | E>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs782438899 CA4299863 |
178 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367724062 rs1554467949 |
180 | Q>* | No |
ClinGen gnomAD |
|
|
rs782277275 CA4299865 |
183 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs587681828 CA4299866 |
183 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4299867 rs782612194 |
184 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA367724115 rs782612194 |
184 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA367724129 rs1307678729 |
185 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA367724145 rs1379283804 |
186 | V>A | No |
ClinGen TOPMed |
|
|
CA4299871 rs781924725 |
187 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782409823 CA367724168 |
188 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782409823 CA4299872 |
188 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1554467959 CA367724199 |
190 | K>T | No |
ClinGen gnomAD |
|
|
rs1368221518 CA367724209 |
191 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA367724216 rs1159850111 |
191 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs202011164 CA4299873 |
192 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs587741041 CA4299874 |
192 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA367724226 rs587741041 |
192 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1451639756 CA367724269 |
196 | G>R | No |
ClinGen TOPMed |
|
|
rs1563129592 CA367724279 |
196 | G>V | No |
ClinGen Ensembl |
|
|
rs1488059286 CA367724307 |
199 | G>S | No |
ClinGen TOPMed gnomAD |
|
| rs1207677864 | 199 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1554467964 CA367724315 |
199 | G>V | No |
ClinGen gnomAD |
|
|
rs1218445650 CA367724325 |
200 | H>R | No |
ClinGen TOPMed |
|
|
CA367724346 rs1554467967 |
202 | R>C | No |
ClinGen gnomAD |
|
|
rs1308391550 CA367724349 |
202 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1239541251 CA367724380 |
205 | V>G | No |
ClinGen TOPMed |
|
|
rs781944432 CA4299880 |
206 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs782689945 CA4299882 |
208 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782689945 CA367724414 |
208 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1554467972 CA367724419 |
209 | D>Y | No |
ClinGen TOPMed |
|
|
rs147290058 COSM1319792 COSM1319793 CA4299883 |
210 | M>T | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA367724434 rs1554467977 |
210 | M>V | No |
ClinGen gnomAD |
|
|
CA4299884 rs782146868 |
216 | Q>* | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 217 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA367724537 rs782766516 |
218 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4299885 rs782766516 |
218 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs141046830 CA4299888 |
219 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4299889 rs141046830 |
219 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4299887 rs587768533 |
219 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA159886728 rs201374916 |
221 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA4299890 rs587639389 |
221 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4299891 rs782662563 |
223 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs1196389434 CA367724608 |
225 | A>V | No |
ClinGen TOPMed |
|
|
rs150827650 CA4299894 |
226 | E>D | No |
ClinGen ESP ExAC gnomAD |
|
|
rs141552293 CA4299893 |
226 | E>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4299896 rs782314081 |
227 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs781918861 CA4299898 |
230 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367724689 rs1487157259 |
231 | Q>E | No |
ClinGen TOPMed |
|
|
rs139250213 CA4299900 |
231 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs150021451 CA367724710 |
232 | F>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4299904 rs781809800 |
233 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782105834 CA4299903 |
233 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs782105834 CA4299902 |
233 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA4299905 rs782057540 |
234 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201856955 CA159886762 |
237 | H>N | No |
ClinGen TOPMed |
|
|
CA367724791 rs201856955 |
237 | H>Y | No |
ClinGen TOPMed |
|
|
rs1584625545 CA367724839 |
239 | E>D | No |
ClinGen Ensembl |
|
|
CA367724830 rs1402270004 |
239 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA4299907 rs781893413 |
241 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA367724893 rs1554468001 |
242 | W>* | No |
ClinGen gnomAD |
|
|
CA4299908 rs139338132 |
242 | W>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1554468039 CA367724996 |
244 | F>I | No |
ClinGen gnomAD |
|
|
rs1554468039 CA367724999 |
244 | F>L | No |
ClinGen gnomAD |
|
|
CA4299922 rs782070085 |
245 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
No associated diseases with Q86UV7
2 GO annotations of cellular component
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| zinc ion binding | Binding to a zinc ion (Zn). |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
49 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E9G4 | TRIM10 | Tripartite motif-containing protein 10 | Bos taurus (Bovine) | PR |
| Q2T9Z0 | TRIM17 | E3 ubiquitin-protein ligase TRIM17 | Bos taurus (Bovine) | PR |
| E1BJS7 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Bos taurus (Bovine) | PR |
| Q7YRV4 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Bos taurus (Bovine) | PR |
| Q1PRL4 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Gallus gallus (Chicken) | PR |
| Q7YR32 | TRIM10 | Tripartite motif-containing protein 10 | Pan troglodytes (Chimpanzee) | PR |
| Q9BTV5 | FSD1 | Fibronectin type III and SPRY domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q9UJV3 | MID2 | Probable E3 ubiquitin-protein ligase MID2 | Homo sapiens (Human) | PR |
| P29590 | PML | Protein PML | Homo sapiens (Human) | PR |
| Q9NQ86 | TRIM36 | E3 ubiquitin-protein ligase TRIM36 | Homo sapiens (Human) | PR |
| Q86UV6 | TRIM74 | Tripartite motif-containing protein 74 | Homo sapiens (Human) | PR |
| Q2Q1W2 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Homo sapiens (Human) | PR |
| Q9BXM9 | FSD1L | FSD1-like protein | Homo sapiens (Human) | PR |
| Q5EBN2 | TRIM61 | Putative tripartite motif-containing protein 61 | Homo sapiens (Human) | PR |
| Q86XT4 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Homo sapiens (Human) | PR |
| Q8N9V2 | TRIML1 | Probable E3 ubiquitin-protein ligase TRIML1 | Homo sapiens (Human) | PR |
| Q9BZY9 | TRIM31 | E3 ubiquitin-protein ligase TRIM31 | Homo sapiens (Human) | PR |
| Q9H2S5 | RNF39 | RING finger protein 39 | Homo sapiens (Human) | PR |
| Q6ZMU5 | TRIM72 | Tripartite motif-containing protein 72 | Homo sapiens (Human) | PR |
| Q9UPQ4 | TRIM35 | E3 ubiquitin-protein ligase TRIM35 | Homo sapiens (Human) | PR |
| Q9C029 | TRIM7 | E3 ubiquitin-protein ligase TRIM7 | Homo sapiens (Human) | PR |
| O15553 | MEFV | Pyrin | Homo sapiens (Human) | SS |
| A6NCK2 | TRIM43B | Tripartite motif-containing protein 43B | Homo sapiens (Human) | PR |
| P19474 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Homo sapiens (Human) | PR |
| Q14142 | TRIM14 | Tripartite motif-containing protein 14 | Homo sapiens (Human) | PR |
| A6NK02 | TRIM75 | Tripartite motif-containing protein 75 | Homo sapiens (Human) | PR |
| Q9WUH5 | Trim10 | Tripartite motif-containing protein 10 | Mus musculus (Mouse) | PR |
| Q8BZT2 | Sh3rf2 | E3 ubiquitin-protein ligase SH3RF2 | Mus musculus (Mouse) | PR |
| Q7TPM3 | Trim17 | E3 ubiquitin-protein ligase TRIM17 | Mus musculus (Mouse) | PR |
| Q1XH17 | Trim72 | Tripartite motif-containing protein 72 | Mus musculus (Mouse) | PR |
| Q60953 | Pml | Protein PML | Mus musculus (Mouse) | PR |
| Q9JJ26 | Mefv | Pyrin | Mus musculus (Mouse) | SS |
| Q99PQ1 | Trim12a | Tripartite motif-containing protein 12A | Mus musculus (Mouse) | PR |
| Q61510 | Trim25 | E3 ubiquitin/ISG15 ligase TRIM25 | Mus musculus (Mouse) | PR |
| Q1PSW8 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Mus musculus (Mouse) | PR |
| Q3TL54 | Trim43a | Tripartite motif-containing protein 43A | Mus musculus (Mouse) | PR |
| P86449 | Trim43c | Tripartite motif-containing protein 43C | Mus musculus (Mouse) | PR |
| Q810I2 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Mus musculus (Mouse) | PR |
| O77666 | TRIM26 | Tripartite motif-containing protein 26 | Sus scrofa (Pig) | PR |
| O19085 | TRIM10 | Tripartite motif-containing protein 10 | Sus scrofa (Pig) | PR |
| Q865W2 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Sus scrofa (Pig) | PR |
| Q920M2 | Rnf39 | RING finger protein 39 | Rattus norvegicus (Rat) | PR |
| Q9JJ25 | Mefv | Pyrin | Rattus norvegicus (Rat) | SS |
| A0JPQ4 | Trim72 | Tripartite motif-containing protein 72 | Rattus norvegicus (Rat) | PR |
| D3ZVM4 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Rattus norvegicus (Rat) | PR |
| Q810I1 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Rattus norvegicus (Rat) | PR |
| F6QEU4 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q640S6 | trim72 | Tripartite motif-containing protein 72 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| E7FAM5 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAWQVSLLEL | EDRLQCPICL | EVFKESLMLQ | CGHSYCKGCL | VSLSYHLDTK | VRCPMCWQVV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DGSSSLPNVS | LAWVIEALRL | PGDPEPKVCV | HHRNPLSLFC | EKDQELICGL | CGLLGSHQHH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PVTPVSTVCS | RMKEELAALF | SELKQEQKKV | DELIAKLVKN | RTRIVNESDV | FSWVIRREFQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ELRHPVDEEK | ARCLEGIGGH | TRGLVASLDM | QLEQAQGTRE | RLAQAECVLE | QFGNEDHHEF |
| IWKFHSMASR |