Q84JX1
Gene name |
PME19 (ARATH5, At1g11590, T23J18.25) |
Protein name |
Probable pectinesterase/pectinesterase inhibitor 19 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G11590 |
EC number |
3.1.1.11: Carboxylic ester hydrolases |
Protein Class |
|
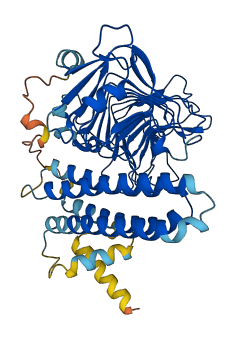
Descriptions
(Annotation from UniProt)
The PMEI region may act as an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
215-509 (Pectinesterase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for Q84JX1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q84JX1-F1 | Predicted | AlphaFoldDB |
179 variants for Q84JX1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH01036524 | 34 | F>L | No | 1000Genomes | |
| ENSVATH01036525 | 38 | L>F | No | 1000Genomes | |
| ENSVATH07509164 | 43 | S>W | No | 1000Genomes | |
| ENSVATH01036529 | 46 | S>L | No | 1000Genomes | |
| ENSVATH04558920 | 61 | S>Y | No | 1000Genomes | |
| ENSVATH10992089 | 67 | M>I | No | 1000Genomes | |
| ENSVATH01036532 | 68 | V>A | No | 1000Genomes | |
| ENSVATH10992090 | 83 | E>D | No | 1000Genomes | |
| tmp_1_3892904_G_A | 91 | E>K | No | 1000Genomes | |
| ENSVATH01036535 | 94 | K>N | No | 1000Genomes | |
| tmp_1_3892920_T_A | 96 | F>Y | No | 1000Genomes | |
| tmp_1_3892936_G_T | 101 | E>D | No | 1000Genomes | |
| tmp_1_3892935_A_G | 101 | E>G | No | 1000Genomes | |
| tmp_1_3892934_G_A | 101 | E>K | No | 1000Genomes | |
| ENSVATH01036536 | 103 | T>A | No | 1000Genomes | |
| ENSVATH10992092 | 111 | V>G | No | 1000Genomes | |
| ENSVATH04558922 | 113 | E>K | No | 1000Genomes | |
| ENSVATH04558923 | 118 | E>Q | No | 1000Genomes | |
| tmp_1_3892991_C_T | 120 | P>S | No | 1000Genomes | |
| ENSVATH01036538 | 122 | R>L | No | 1000Genomes | |
| tmp_1_3893000_G_A | 123 | G>R | No | 1000Genomes | |
| ENSVATH13880674 | 125 | Y>C | No | 1000Genomes | |
| ENSVATH01036539 | 126 | E>Q | No | 1000Genomes | |
| ENSVATH13880695 | 128 | V>A | No | 1000Genomes | |
| tmp_1_3893026_G_A | 131 | W>* | No | 1000Genomes | |
| ENSVATH07509169 | 133 | S>N | No | 1000Genomes | |
| ENSVATH07509169 | 133 | S>T | No | 1000Genomes | |
| tmp_1_3893039_C_T | 136 | L>F | No | 1000Genomes | |
| ENSVATH07509170 | 137 | T>I | No | 1000Genomes | |
| tmp_1_3893078_G_A | 149 | G>S | No | 1000Genomes | |
| ENSVATH10992094 | 149 | G>V | No | 1000Genomes | |
| tmp_1_3893090_C_T | 153 | R>C | No | 1000Genomes | |
| ENSVATH04558927 | 157 | P>L | No | 1000Genomes | |
| ENSVATH04558928 | 160 | E>K | No | 1000Genomes | |
| ENSVATH13880697 | 161 | D>H | No | 1000Genomes | |
| ENSVATH13880697 | 161 | D>N | No | 1000Genomes | |
| tmp_1_3893117_C_T | 162 | L>F | No | 1000Genomes | |
| ENSVATH04558930 | 168 | I>V | No | 1000Genomes | |
| tmp_1_3893139_CT_GT,AT,C | 169 | A>D | No | 1000Genomes | |
| tmp_1_3893139_CT_GT,AT,C | 169 | A>G | No | 1000Genomes | |
| ENSVATH01036543 | 171 | A>T | No | 1000Genomes | |
| ENSVATH13880698 | 177 | S>L | No | 1000Genomes | |
| tmp_1_3893169_G_A | 179 | R>K | No | 1000Genomes | |
| ENSVATH01036545 | 187 | V>A | No | 1000Genomes | |
| ENSVATH01036544 | 187 | V>M | No | 1000Genomes | |
| ENSVATH04558934 | 189 | P>S | No | 1000Genomes | |
| ENSVATH10992284 | 191 | S>G | No | 1000Genomes | |
| tmp_1_3893205_G_T | 191 | S>I | No | 1000Genomes | |
| ENSVATH10992435 | 192 | P>S | No | 1000Genomes | |
| tmp_1_3893211_C_T | 193 | S>F | No | 1000Genomes | |
| tmp_1_3893215_G_A | 194 | W>* | No | 1000Genomes | |
| ENSVATH10992436 | 195 | L>S | No | 1000Genomes | |
| ENSVATH10992437 | 197 | H>P | No | 1000Genomes | |
| tmp_1_3893228_G_C | 199 | D>H | No | 1000Genomes | |
| tmp_1_3893237_G_A | 202 | D>N | No | 1000Genomes | |
| ENSVATH07509175 | 207 | A>T | No | 1000Genomes | |
| ENSVATH04558936 | 208 | E>K | No | 1000Genomes | |
| ENSVATH07509194 | 209 | A>T | No | 1000Genomes | |
| ENSVATH13880702 | 209 | A>V | No | 1000Genomes | |
| ENSVATH01036554 | 214 | A>D | No | 1000Genomes | |
| ENSVATH04558952 | 214 | A>P | No | 1000Genomes | |
| ENSVATH01036554 | 214 | A>V | No | 1000Genomes | |
| tmp_1_3893584_G_T | 216 | V>F | No | 1000Genomes | |
| ENSVATH01036555 | 222 | G>E | No | 1000Genomes | |
| ENSVATH01036558 | 236 | A>T | No | 1000Genomes | |
| ENSVATH01036560 | 243 | R>K | No | 1000Genomes | |
| ENSVATH04558957 | 257 | V>I | No | 1000Genomes | |
| tmp_1_3893729_C_T | 264 | P>L | No | 1000Genomes | |
| ENSVATH10992601 | 264 | P>S | No | 1000Genomes | |
| tmp_1_3893740_C_A | 268 | L>I | No | 1000Genomes | |
| tmp_1_3893746_G_C | 270 | G>R | No | 1000Genomes | |
| tmp_1_3893753_G_A | 272 | G>D | No | 1000Genomes | |
| ENSVATH10992602 | 275 | L>S | No | 1000Genomes | |
| ENSVATH01036561 | 280 | S>G | No | 1000Genomes | |
| ENSVATH04558958 | 284 | A>S | No | 1000Genomes | |
| ENSVATH01036562 | 285 | S>T | No | 1000Genomes | |
| tmp_1_3893794_A_C | 286 | N>H | No | 1000Genomes | |
| tmp_1_3893813_A_G | 292 | N>S | No | 1000Genomes | |
| tmp_1_3893816_C_T | 293 | T>I | No | 1000Genomes | |
| tmp_1_3893819_C_T | 294 | A>V | No | 1000Genomes | |
| ENSVATH10992604 | 295 | T>I | No | 1000Genomes | |
| ENSVATH04558959 | 296 | V>I | No | 1000Genomes | |
| ENSVATH13880717 | 305 | G>V | No | 1000Genomes | |
| ENSVATH10992660 | 314 | A>V | No | 1000Genomes | |
| tmp_1_3893994_G_A | 317 | A>T | No | 1000Genomes | |
| tmp_1_3894012_G_A | 323 | A>T | No | 1000Genomes | |
| ENSVATH10992662 | 325 | R>C | No | 1000Genomes | |
| ENSVATH10992663 | 328 | G>C | No | 1000Genomes | |
| ENSVATH10992663 | 328 | G>S | No | 1000Genomes | |
| ENSVATH01036570 | 344 | A>T | No | 1000Genomes | |
| ENSVATH10992664 | 344 | A>V | No | 1000Genomes | |
| ENSVATH10992716 | 351 | R>C | No | 1000Genomes | |
| tmp_1_3894105_T_A | 354 | Y>N | No | 1000Genomes | |
| ENSVATH04558964 | 362 | T>I | No | 1000Genomes | |
| ENSVATH10992717 | 366 | I>L | No | 1000Genomes | |
| tmp_1_3894148_G_C | 368 | G>A | No | 1000Genomes | |
| tmp_1_3894157_T_C | 371 | V>A | No | 1000Genomes | |
| ENSVATH04558965 | 372 | A>T | No | 1000Genomes | |
| ENSVATH10992718 | 372 | A>V | No | 1000Genomes | |
| tmp_1_3894174_T_A | 377 | C>S | No | 1000Genomes | |
| tmp_1_3894175_GC_G,AC | 377 | C>Y | No | 1000Genomes | |
| tmp_1_3894187_C_T | 381 | A>V | No | 1000Genomes | |
| ENSVATH10992722 | 383 | Q>* | No | 1000Genomes | |
| ENSVATH04558967 | 384 | P>A | No | 1000Genomes | |
| tmp_1_3894205_G_A | 387 | G>E | No | 1000Genomes | |
| ENSVATH01036572 | 388 | Q>* | No | 1000Genomes | |
| tmp_1_3894219_A_C | 392 | I>L | No | 1000Genomes | |
| tmp_1_3894237_G_A | 398 | A>T | No | 1000Genomes | |
| tmp_1_3894240_T_A | 399 | F>I | No | 1000Genomes | |
| tmp_1_3894241_T_A | 399 | F>Y | No | 1000Genomes | |
| tmp_1_3894250_T_A | 402 | I>N | No | 1000Genomes | |
| tmp_1_3894249_A_G | 402 | I>V | No | 1000Genomes | |
| tmp_1_3894252_T_A | 403 | Y>N | No | 1000Genomes | |
| tmp_1_3894259_G_A | 405 | G>D | No | 1000Genomes | |
| ENSVATH10992724 | 405 | G>S | No | 1000Genomes | |
| tmp_1_3894263_C_A | 406 | F>L | No | 1000Genomes | |
| tmp_1_3894264_A_T | 407 | T>S | No | 1000Genomes | |
| tmp_1_3894286_C_G | 414 | T>S | No | 1000Genomes | |
| tmp_1_3894289_C_G | 415 | A>G | No | 1000Genomes | |
| ENSVATH04558970 | 417 | S>L | No | 1000Genomes | |
| tmp_1_3894298_A_T | 418 | D>V | No | 1000Genomes | |
| tmp_1_3894325_A_G | 427 | Y>C | No | 1000Genomes | |
| tmp_1_3894328_T_G | 428 | L>R | No | 1000Genomes | |
| tmp_1_3894331_GA_AA,G | 429 | G>E | No | 1000Genomes | |
| tmp_1_3894337_C_T | 431 | P>L | No | 1000Genomes | |
| ENSVATH04558975 | 432 | W>* | No | 1000Genomes | |
| ENSVATH04558976 | 433 | R>K | No | 1000Genomes | |
| tmp_1_3894352_C_T | 436 | S>L | No | 1000Genomes | |
| ENSVATH10992796 | 438 | V>I | No | 1000Genomes | |
| ENSVATH01036573 | 440 | V>I | No | 1000Genomes | |
| tmp_1_3894368_G_A | 441 | M>I | No | 1000Genomes | |
| ENSVATH04558977 | 443 | S>C | No | 1000Genomes | |
| ENSVATH04558977 | 443 | S>F | No | 1000Genomes | |
| ENSVATH04558978 | 446 | G>D | No | 1000Genomes | |
| tmp_1_3894384_G_A | 447 | D>N | No | 1000Genomes | |
| tmp_1_3894393_G_A | 450 | D>N | No | 1000Genomes | |
| tmp_1_3894397_C_T | 451 | P>L | No | 1000Genomes | |
| ENSVATH10992797 | 451 | P>S | No | 1000Genomes | |
| ENSVATH04558979 | 452 | A>P | No | 1000Genomes | |
| ENSVATH04558980 | 452 | A>V | No | 1000Genomes | |
| ENSVATH10992799 | 453 | G>S | No | 1000Genomes | |
| tmp_1_3894406_G_T | 454 | W>L | No | 1000Genomes | |
| ENSVATH01036575 | 455 | T>I | No | 1000Genomes | |
| ENSVATH01036575 | 455 | T>S | No | 1000Genomes | |
| ENSVATH04558982 | 456 | P>L | No | 1000Genomes | |
| ENSVATH13880721 | 456 | P>S | No | 1000Genomes | |
| tmp_1_3894416_G_A | 457 | W>* | No | 1000Genomes | |
| tmp_1_3894415_G_A | 457 | W>* | No | 1000Genomes | |
| tmp_1_3894417_G_A | 458 | E>K | No | 1000Genomes | |
| tmp_1_3894420_G_A | 459 | G>R | No | 1000Genomes | |
| tmp_1_3894423_G_A | 460 | E>K | No | 1000Genomes | |
| tmp_1_3894429_G_T,A | 462 | G>C | No | 1000Genomes | |
| tmp_1_3894430_G_A | 462 | G>D | No | 1000Genomes | |
| tmp_1_3894429_G_T,A | 462 | G>S | No | 1000Genomes | |
| ENSVATH04558983 | 464 | S>L | No | 1000Genomes | |
| ENSVATH13880722 | 466 | L>F | No | 1000Genomes | |
| ENSVATH04558984 | 467 | H>Y | No | 1000Genomes | |
| ENSVATH04558985 | 469 | R>C | No | 1000Genomes | |
| ENSVATH10992800 | 469 | R>H | No | 1000Genomes | |
| ENSVATH10992801 | 470 | E>K | No | 1000Genomes | |
| ENSVATH04558986 | 474 | R>K | No | 1000Genomes | |
| ENSVATH04558987 | 476 | P>L | No | 1000Genomes | |
| ENSVATH01036577 | 477 | G>R | No | 1000Genomes | |
| ENSVATH01036578 | 478 | A>D | No | 1000Genomes | |
| ENSVATH01036578 | 478 | A>G | No | 1000Genomes | |
| ENSVATH01036578 | 478 | A>V | No | 1000Genomes | |
| ENSVATH01036580 | 483 | R>G | No | 1000Genomes | |
| ENSVATH04558988 | 483 | R>K | No | 1000Genomes | |
| tmp_1_3894502_G_A | 486 | W>* | No | 1000Genomes | |
| ENSVATH10992803 | 488 | G>S | No | 1000Genomes | |
| tmp_1_3894513_A_T | 490 | K>* | No | 1000Genomes | |
| ENSVATH01036582 | 494 | D>N | No | 1000Genomes | |
| ENSVATH01036582 | 494 | D>Y | No | 1000Genomes | |
| ENSVATH04558991 | 500 | E>K | No | 1000Genomes | |
| ENSVATH04558991 | 500 | E>Q | No | 1000Genomes | |
| ENSVATH10992876 | 503 | V>L | No | 1000Genomes | |
| ENSVATH04558992 | 504 | A>T | No | 1000Genomes | |
| tmp_1_3894567_G_T | 508 | D>Y | No | 1000Genomes | |
| ENSVATH04558993 | 509 | G>E | No | 1000Genomes |
No associated diseases with Q84JX1
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.1.11 | Carboxylic ester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| aspartyl esterase activity | Catalysis of the hydrolysis of an ester bond by a mechanism involving a catalytically active aspartic acid residue. |
| pectinesterase activity | Catalysis of the reaction: pectin + n H2O = n methanol + pectate. |
| pectinesterase inhibitor activity | Binds to and stops, prevents or reduces the activity of pectinesterase. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall modification | The series of events leading to chemical and structural alterations of an existing cell wall that can result in loosening, increased extensibility or disassembly. |
| pectin catabolic process | The chemical reactions and pathways resulting in the breakdown of pectin, a polymer containing a backbone of alpha-1,4-linked D-galacturonic acid residues. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q1JPL7 | PME18 | Pectinesterase/pectinesterase inhibitor 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1PEC0 | PME42 | Probable pectinesterase/pectinesterase inhibitor 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SRX4 | PME7 | Probable pectinesterase/pectinesterase inhibitor 7 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49298 | PME6 | Probable pectinesterase/pectinesterase inhibitor 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42534 | PME2 | Pectinesterase 2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q43867 | PME1 | Pectinesterase 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O48711 | PME12 | Probable pectinesterase/pectinesterase inhibitor 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7Y201 | PME13 | Probable pectinesterase/pectinesterase inhibitor 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SKX2 | PME16 | Probable pectinesterase/pectinesterase inhibitor 16 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22149 | PME17 | Probable pectinesterase/pectinesterase inhibitor 17 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O80722 | PME4 | Pectinesterase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV8 | PME5 | Pectinesterase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22256 | PME20 | Probable pectinesterase/pectinesterase inhibitor 20 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GX86 | PME21 | Probable pectinesterase/pectinesterase inhibitor 21 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M9W7 | PME22 | Putative pectinesterase/pectinesterase inhibitor 22 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GXA1 | PME23 | Probable pectinesterase/pectinesterase inhibitor 23 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SG77 | PME24 | Putative pectinesterase/pectinesterase inhibitor 24 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49006 | PME3 | Pectinesterase/pectinesterase inhibitor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9STY3 | PME33 | Probable pectinesterase/pectinesterase inhibitor 33 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LYT5 | PME35 | Probable pectinesterase/pectinesterase inhibitor 35 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84R10 | PME36 | Probable pectinesterase/pectinesterase inhibitor 36 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV6 | VGDH2 | Probable pectinesterase/pectinesterase inhibitor VGDH2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81320 | PME38 | Putative pectinesterase/pectinesterase inhibitor 38 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81415 | PME39 | Probable pectinesterase/pectinesterase inhibitor 39 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81301 | PME40 | Probable pectinesterase/pectinesterase inhibitor 40 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8RXK7 | PME41 | Probable pectinesterase/pectinesterase inhibitor 41 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O23447 | PME43 | Putative pectinesterase/pectinesterase inhibitor 43 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY7 | PME44 | Probable pectinesterase/pectinesterase inhibitor 44 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY6 | PME45 | Putative pectinesterase/pectinesterase inhibitor 45 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF78 | PME46 | Probable pectinesterase/pectinesterase inhibitor 46 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF77 | PME47 | Probable pectinesterase/pectinesterase inhibitor 47 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXD9 | PME51 | Probable pectinesterase/pectinesterase inhibitor 51 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E989 | PME54 | Probable pectinesterase/pectinesterase inhibitor 54 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E8Z8 | PME28 | Putative pectinesterase/pectinesterase inhibitor 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FJ21 | PME58 | Probable pectinesterase/pectinesterase inhibitor 58 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN5 | PME59 | Probable pectinesterase/pectinesterase inhibitor 59 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN4 | PME60 | Probable pectinesterase/pectinesterase inhibitor 60 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FK05 | PME61 | Probable pectinesterase/pectinesterase inhibitor 61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8L7Q7 | PME64 | Probable pectinesterase/pectinesterase inhibitor 64 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q94CB1 | PME25 | Probable pectinesterase/pectinesterase inhibitor 25 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXK7 | PME32 | Probable pectinesterase/pectinesterase inhibitor 32 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M3B0 | PME34 | Probable pectinesterase/pectinesterase inhibitor 34 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P09607 | PME2.1 | Pectinesterase 2.1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| P14280 | PME1.9 | Pectinesterase 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96576 | PME3 | Pectinesterase 3 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96575 | PME2.2 | Pectinesterase 2.2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLVKVFSFFI | LMITMVVIGV | SKEYCDDKQS | CQNFLLELKA | GSSSLSEIRR | RDLLIIVLKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SVRRIDMVMI | GVMDDTKQHE | EMENDLLGVK | EDTKLFEEMM | ESTKDRMIRS | VEELLGGEFP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NRGSYENVHT | WLSSVLTSYI | TCIDEIGEGA | YKRRVEPKLE | DLISRARIAL | ALFISISPRD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NTELISVIPN | SPSWLFHVDK | KDLYLNAEAL | KKIADVVVAK | DGTGKYSTVN | AAIAAAPQHS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QKRFVIYIKT | GIYDEIVVIE | NTKPNLTLIG | DGQDLTIITS | NLSASNVRRT | FNTATVASNG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NGFIGVDMCF | RNTAGPAKGP | AVALRVSGDM | SVIYRCRVEG | YQDALYPHSD | RQFYRECFIT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GTVDFICGNA | VAVFQFCQIV | ARQPKMGQSN | VITAQSRAFK | DIYSGFTIQK | CNITASSDLD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TTTVKTYLGR | PWRIFSTVAV | MQSFIGDLVD | PAGWTPWEGE | TGLSTLHYRE | YQNRGPGAVT |
| 490 | 500 | 510 | 520 | ||
| SRRVKWSGFK | VMKDPKQATE | FTVAKLLDGE | TWLKETRIPY | ESGL |