Q83RJ4
Gene name |
ipaH3 (SF1383) |
Protein name |
E3 ubiquitin-protein ligase ipaH3 |
Names |
RING-type E3 ubiquitin transferase ipaH3 |
Species |
Shigella flexneri |
KEGG Pathway |
sfl:SF1383 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
FAMILY NOT NAMED (PTHR47114) |
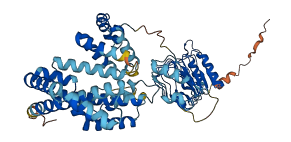
Descriptions
The IpaH family of novel E3 ligase (NEL) enzymes occur in a variety of pathogenic and commensal bacteria that interact with eukaryotic hosts. The leucine-rich repeat (LRR) substrate recognition domains of different IpaH enzymes autoinhibit the enzymatic activity of the adjacent catalytic novel E3 ligase domain. Removal of the LRR domain increases autoubiquitination and potentiates free polyubiquitin chain synthesis.
Autoinhibitory domains (AIDs)
Target domain |
287-497 (E3 ligase domain) |
Relief mechanism |
|
Assay |
Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q83RJ4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3CVR | X-ray | 280 A | A | 1-571 | PDB |
| AF-Q83RJ4-F1 | Predicted | AlphaFoldDB |
No variants for Q83RJ4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q83RJ4 | |||||
No associated diseases with Q83RJ4
4 regional properties for Q83RJ4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 121 - 142 | IPR001611-1 |
| repeat | Leucine-rich repeat | 161 - 182 | IPR001611-2 |
| domain | Novel E3 ligase domain | 287 - 497 | IPR029487 |
| domain | LRR-containing bacterial E3 ligase, N-terminal | 7 - 51 | IPR032674 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47114 | FAMILY NOT NAMED |
| PANTHER Subfamily | PTHR47114:SF2 | OLIGODENDROCYTE-MYELIN GLYCOPROTEIN |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| host cell cytoplasm | The cytoplasm of a host cell. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSIMLPINNN | FSLSQNSFYN | TISGTYADYF | SAWDKWEKQA | LPGENRNEAV | SLLKECLINQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FSELQLNRLN | LSSLPDNLPP | QITVLEITQN | ALISLPELPA | SLEYLDACDN | RLSTLPELPA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SLKHLDVDNN | QLTMLPELPA | LLEYINADNN | QLTMLPELPT | SLEVLSVRNN | QLTFLPELPE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SLEALDVSTN | LLESLPAVPV | RNHHSEETEI | FFRCRENRIT | HIPENILSLD | PTCTIILEDN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PLSSRIRESL | SQQTAQPDYH | GPRIYFSMSD | GQQNTLHRPL | ADAVTAWFPE | NKQSDVSQIW |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HAFEHEEHAN | TFSAFLDRLS | DTVSARNTSG | FREQVAAWLE | KLSTSAELRQ | QSFAVAADAT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ESCEDRVALT | WNNLRKTLLV | HQASEGLFDN | DTGALLSLGR | EMFRLEILED | IARDKVRTLH |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FVDEIEVYLA | FQTMLAEKLQ | LSTAVKEMRF | YGVSGVTAND | LRTAEAMVRS | REENEFTDWF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SLWGPWHAVL | KRTEADRWAQ | AEEQKYEMLE | NEYSQRVADR | LKASGLSGDA | DAEREAGAQV |
| 550 | 560 | 570 | |||
| MRETEQQIYR | QVTDEVLALR | LSENGSQLHH | S |