Q80XA6
Gene name |
Reps2 (Pob1) |
Protein name |
RalBP1-associated Eps domain-containing protein 2 |
Names |
Partner of RalBP1, RalBP1-interacting protein 2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:194590 |
EC number |
|
Protein Class |
|
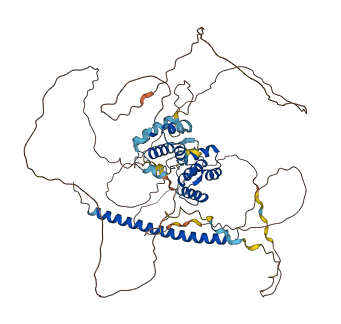
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q80XA6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q80XA6-F1 | Predicted | AlphaFoldDB |
35 variants for Q80XA6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389582702 | 9 | C>Y | No | EVA | |
| rs3411257499 | 74 | E>D | No | EVA | |
| rs3411144418 | 79 | I>N | No | EVA | |
| rs3412351673 | 80 | T>I | No | EVA | |
| rs3411144404 | 81 | E>K | No | EVA | |
| rs3389575545 | 82 | L>Q | No | EVA | |
| rs3389540503 | 83 | C>R | No | EVA | |
| rs3412893572 | 87 | R>* | No | EVA | |
| rs3389578223 | 91 | F>L | No | EVA | |
| rs3389489259 | 106 | A>E | No | EVA | |
| rs3389540441 | 109 | G>S | No | EVA | |
| rs3389585916 | 133 | G>R | No | EVA | |
| rs244925554 | 175 | T>A | No | EVA | |
| rs229218000 | 175 | T>R | No | EVA | |
| rs211983755 | 198 | Y>S | No | EVA | |
| rs241139493 | 221 | P>S | No | EVA | |
| rs260008700 | 228 | G>D | No | EVA | |
| rs243225974 | 228 | G>S | No | EVA | |
| rs3389582672 | 367 | A>G | No | EVA | |
| rs260595227 | 397 | A>T | No | EVA | |
| rs1134124412 | 496 | S>P | No | EVA | |
| rs1131701370 | 497 | G>E | No | EVA | |
| rs31346967 | 524 | S>P | No | EVA | |
| rs31346964 | 541 | M>K | No | EVA | |
| rs3409837519 | 543 | A>S | No | EVA | |
| rs3412441928 | 547 | Q>L | No | EVA | |
| rs3412643697 | 576 | A>T | No | EVA | |
| rs214519010 | 577 | V>A | No | EVA | |
| rs3411141617 | 581 | M>V | No | EVA | |
| rs3412126807 | 593 | S>* | No | EVA | |
| rs3412528937 | 595 | Q>P | No | EVA | |
| rs3412439334 | 596 | K>N | No | EVA | |
| rs3409837562 | 599 | I>L | No | EVA | |
| rs3411252224 | 600 | Q>R | No | EVA | |
| rs257535486 | 606 | N>H | No | EVA |
No associated diseases with Q80XA6
4 regional properties for Q80XA6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | EH domain | 21 - 122 | IPR000261-1 |
| domain | EH domain | 261 - 356 | IPR000261-2 |
| domain | EF-hand domain | 301 - 336 | IPR002048 |
| binding_site | EF-Hand 1, calcium-binding site | 314 - 326 | IPR018247 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| endocytosis | A vesicle-mediated transport process in which cells take up external materials or membrane constituents by the invagination of a small region of the plasma membrane to form a new membrane-bounded vesicle. |
| endosomal transport | The directed movement of substances mediated by an endosome, a membrane-bounded organelle that carries materials enclosed in the lumen or located in the endosomal membrane. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEVAAGGGCG | AGPPLLLSDS | EQQCYSELFA | RCAGAASGGP | GPGPPEATRV | APGTATAAAG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PVADLFRASQ | LPPETLHQIT | ELCGAKRVGY | FGPTQFYVAL | KLIAAAQAGL | PVRTESIKCE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LPLPRFVMSK | NDGEIRFGNP | AELHGPKVQI | PYLTTEKNSF | KRMDNEDKQE | TQSPTMSPLA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SPPSSPPHYQ | RVSLSHGYSK | LRSGTEQMHP | APYERQPIGQ | PEGPSSEGPG | AKPFRRQASL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IRSFSVEREP | QENNSNYPDE | PWRITEEQRE | YYVNQFRSLQ | PDPSSFISGS | VAKNFFTKSK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LSIPELSYIW | ELSDADCDGA | LTLSEFCAAF | HLIVARKNGY | PLPEGLPPTL | QPEYLQAAFP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KSKWECAIFD | SYSESMPANQ | QSCDLNRMEK | TSVKDVADFP | VPTQDVTTAD | DKQALKSTVN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ESLPKDVSED | TATSKDYNSL | KARPRSRSYS | STSIEEAMKR | GEDPPTPPPR | PQKTHSRASS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| LDLNKVFLPS | APAANSGLLP | PPPALPPRPC | PTQSEPVSEA | DLHSQLNRAP | SQAAESSPTK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| MDAPHAQPPS | KPIRRKFRPE | NQTTESQEPA | AAVGGAVSAA | MVKPHPTVQK | QSSKQKKAIQ |
| 610 | 620 | 630 | 640 | ||
| TAIRKNKEAN | AVLARLNSEL | QQQLKEVHQE | RIALENQLEQ | LRPVTVL |