Q80WG5
Gene name |
Lrrc8a |
Protein name |
Volume-regulated anion channel subunit LRRC8A |
Names |
Leucine-rich repeat-containing protein 8A, Protein ebouriffe, ebo |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:241296 |
EC number |
|
Protein Class |
|
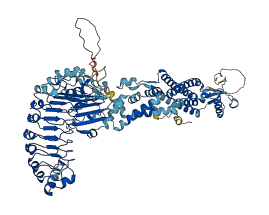
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

32 structures for Q80WG5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6FNW | X-ray | 180 A | A | 398-810 | PDB |
| 6G8Z | EM | 366 A | A/B/C/D/E/F | 1-810 | PDB |
| 6G9L | EM | 501 A | A/B/C/D/E/F | 1-810 | PDB |
| 6G9O | EM | 425 A | A/B/C/D/E/F | 1-810 | PDB |
| 6NZW | EM | 321 A | A/B/C/D/E/F | 1-810 | PDB |
| 6NZZ | EM | 332 A | A/B/C/D/E/F | 1-810 | PDB |
| 6O00 | EM | 418 A | A/B/C/D/E/F | 1-810 | PDB |
| 7M17 | EM | 365 A | A/B/C/D/E/F | 1-810 | PDB |
| 7M19 | EM | 369 A | A/B/C/D/E/F | 1-810 | PDB |
| 7P60 | EM | 380 A | A/B/C/D/E/F | 1-810 | PDB |
| 7P6K | EM | 380 A | A/B/C/D/E/F | 1-810 | PDB |
| 8B41 | EM | 380 A | A/B/C/D | 2-810 | PDB |
| 8B42 | EM | 660 A | A/B/C/D | 2-810 | PDB |
| 8DR8 | EM | 304 A | PDB | ||
| 8DRA | EM | 398 A | PDB | ||
| 8DRE | EM | 318 A | PDB | ||
| 8DRK | EM | 295 A | PDB | ||
| 8DRN | EM | 412 A | PDB | ||
| 8DRO | EM | 406 A | PDB | ||
| 8DRQ | EM | 416 A | PDB | ||
| 8DS3 | EM | 307 A | PDB | ||
| 8DS9 | EM | 317 A | PDB | ||
| 8DSA | EM | 348 A | PDB | ||
| 8F74 | EM | 310 A | PDB | ||
| 8F75 | EM | 400 A | A/B | 91-810 | PDB |
| 8F77 | EM | 315 A | PDB | ||
| 8F79 | EM | 315 A | PDB | ||
| 8F7B | EM | 315 A | PDB | ||
| 8F7D | EM | 420 A | PDB | ||
| 8F7E | EM | 413 A | PDB | ||
| 8F7J | EM | 432 A | PDB | ||
| AF-Q80WG5-F1 | Predicted | AlphaFoldDB |
37 variants for Q80WG5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388537032 | 7 | L>F | No | EVA | |
| rs3388539640 | 39 | A>D | No | EVA | |
| rs3388540997 | 40 | V>D | No | EVA | |
| rs3388534490 | 58 | K>N | No | EVA | |
| rs3388543714 | 66 | N>S | No | EVA | |
| rs3388545011 | 69 | F>L | No | EVA | |
| rs3391407999 | 72 | W>* | No | EVA | |
| rs3388542446 | 93 | G>R | No | EVA | |
| rs3388543784 | 127 | Y>F | No | EVA | |
| rs864284750 | 127 | Y>H | No | EVA | |
| rs864284750 | 127 | Y>N | No | EVA | |
| rs3388544908 | 158 | S>F | No | EVA | |
| rs3388537006 | 163 | C>F | No | EVA | |
| rs3388539621 | 180 | E>V | No | EVA | |
| rs3388544974 | 243 | L>M | No | EVA | |
| rs3388543709 | 262 | R>S | No | EVA | |
| rs3388539330 | 278 | I>F | No | EVA | |
| rs3388539639 | 313 | P>A | No | EVA | |
| rs3388542492 | 346 | R>C | No | EVA | |
| rs3388537019 | 353 | F>L | No | EVA | |
| rs3388542818 | 372 | F>L | No | EVA | |
| rs3388539349 | 374 | F>L | No | EVA | |
| rs3388542422 | 429 | E>V | No | EVA | |
| 443 | F>del | ebo; causes male sterility [UniProt] | No | ||
| rs3388539285 | 463 | P>S | No | EVA | |
| rs3388542329 | 475 | W>R | No | EVA | |
| rs3388541044 | 523 | L>M | No | EVA | |
| rs3388544315 | 615 | N>D | No | EVA | |
| rs3388541057 | 649 | N>S | No | EVA | |
| rs3388542456 | 671 | R>G | No | EVA | |
| rs3391422024 | 702 | A>V | No | EVA | |
| rs3388545017 | 706 | L>I | No | EVA | |
| rs27209217 | 715 | V>I | No | EVA | |
| rs3388541958 | 728 | F>L | No | EVA | |
| rs3388539617 | 774 | G>R | No | EVA | |
| rs3388539656 | 780 | K>T | No | EVA | |
| rs3388542351 | 809 | Q>* | No | EVA |
1 associated diseases with Q80WG5
Without disease ID
12 regional properties for Q80WG5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 592 - 651 | IPR001611-1 |
| repeat | Leucine-rich repeat | 662 - 707 | IPR001611-2 |
| repeat | Leucine-rich repeat | 709 - 766 | IPR001611-3 |
| repeat | Leucine-rich repeat, typical subtype | 590 - 613 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 614 - 636 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 638 - 660 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 661 - 684 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 685 - 706 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 707 - 730 | IPR003591-6 |
| repeat | Leucine-rich repeat, typical subtype | 731 - 751 | IPR003591-7 |
| repeat | Leucine-rich repeat, typical subtype | 753 - 776 | IPR003591-8 |
| domain | LRRC8, pannexin-like TM region | 1 - 340 | IPR021040 |
Functions
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell surface | The external part of the cell wall and/or plasma membrane. |
| integral component of lysosomal membrane | The component of the lysosome membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| ion channel complex | A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| anion channel activity | Enables the energy-independent passage of anions across a lipid bilayer down a concentration gradient. |
| cyclic-GMP-AMP transmembrane transporter activity | Enables the transfer of cyclic-GMP-AMP from one side of a membrane to the other. |
| identical protein binding | Binding to an identical protein or proteins. |
| volume-sensitive anion channel activity | Enables the transmembrane transfer of an anion by a volume-sensitive channel. An anion is a negatively charged ion. A volume-sensitive channel is a channel that responds to changes in the volume of a cell. |
14 GO annotations of biological process
| Name | Definition |
|---|---|
| anion transmembrane transport | The process in which an anion is transported across a membrane. |
| anion transport | The directed movement of anions, atoms or small molecules with a net negative charge, into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| aspartate transmembrane transport | The process in which aspartate is transported across a lipid bilayer, from one side of a membrane to the other. |
| cell volume homeostasis | Any process involved in maintaining the steady state of a cell's volume. The cell's volume refers to the three-dimensional space occupied by a cell. |
| cellular glucose homeostasis | A cellular homeostatic process involved in the maintenance of an internal steady state of glucose within a cell or between a cell and its external environment. |
| chloride transmembrane transport | The process in which chloride is transported across a membrane. |
| cyclic-GMP-AMP transmembrane import across plasma membrane | The directed movement of cyclic-GMP-AMP from outside of a cell, across the plasma membrane and into the cytosol. |
| positive regulation of insulin secretion | Any process that activates or increases the frequency, rate or extent of the regulated release of insulin. |
| positive regulation of myoblast differentiation | Any process that activates or increases the frequency, rate or extent of myoblast differentiation. A myoblast is a mononucleate cell type that, by fusion with other myoblasts, gives rise to the myotubes that eventually develop into skeletal muscle fibers. |
| pre-B cell differentiation | The process in which a precursor cell type acquires the specialized features of a pre-B cell. Pre-B cells follow the pro-B cell stage of immature B cell differentiation and undergo rearrangement of heavy chain V, D, and J gene segments. |
| protein hexamerization | The formation of a protein hexamer, a macromolecular structure consisting of six noncovalently associated identical or nonidentical subunits. |
| response to osmotic stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating an increase or decrease in the concentration of solutes outside the organism or cell. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
| taurine transport | The directed movement of taurine into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
36 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3MHH9 | ECM2 | Extracellular matrix protein 2 | Bos taurus (Bovine) | PR |
| P58874 | OPTC | Opticin | Bos taurus (Bovine) | PR |
| Q24K06 | LRRC10 | Leucine-rich repeat-containing protein 10 | Bos taurus (Bovine) | PR |
| Q9V780 | Lap1 | Protein lap1 | Drosophila melanogaster (Fruit fly) | PR |
| Q96NW7 | LRRC7 | Leucine-rich repeat-containing protein 7 | Homo sapiens (Human) | PR |
| Q9HCJ2 | LRRC4C | Leucine-rich repeat-containing protein 4C | Homo sapiens (Human) | PR |
| Q9UFC0 | LRWD1 | Leucine-rich repeat and WD repeat-containing protein 1 | Homo sapiens (Human) | PR |
| Q96L50 | LRR1 | Leucine-rich repeat protein 1 | Homo sapiens (Human) | PR |
| Q86UN2 | RTN4RL1 | Reticulon-4 receptor-like 1 | Homo sapiens (Human) | PR |
| Q8IWK6 | ADGRA3 | Adhesion G protein-coupled receptor A3 | Homo sapiens (Human) | PR |
| Q96FE5 | LINGO1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| Q7L1W4 | LRRC8D | Volume-regulated anion channel subunit LRRC8D | Homo sapiens (Human) | PR |
| Q8TDW0 | LRRC8C | Volume-regulated anion channel subunit LRRC8C | Homo sapiens (Human) | PR |
| Q8IWT6 | LRRC8A | Volume-regulated anion channel subunit LRRC8A | Homo sapiens (Human) | PR |
| Q7TT36 | Adgra3 | Adhesion G protein-coupled receptor A3 | Mus musculus (Mouse) | PR |
| P59383 | Lrrn4 | Leucine-rich repeat neuronal protein 4 | Mus musculus (Mouse) | PR |
| Q9D1T0 | Lingo1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Mus musculus (Mouse) | PR |
| Q8K0S5 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Mus musculus (Mouse) | PR |
| Q80TE7 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Mus musculus (Mouse) | PR |
| Q5RKR3 | Islr2 | Immunoglobulin superfamily containing leucine-rich repeat protein 2 | Mus musculus (Mouse) | PR |
| Q8C031 | Lrrc4c | Leucine-rich repeat-containing protein 4C | Mus musculus (Mouse) | PR |
| Q8BGI7 | Lrrc39 | Leucine-rich repeat-containing protein 39 | Mus musculus (Mouse) | PR |
| Q8R502 | Lrrc8c | Volume-regulated anion channel subunit LRRC8C | Mus musculus (Mouse) | PR |
| Q9D9Q0 | Lrrc69 | Leucine-rich repeat-containing protein 69 | Mus musculus (Mouse) | PR |
| A6H694 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Mus musculus (Mouse) | PR |
| Q5DU41 | Lrrc8b | Volume-regulated anion channel subunit LRRC8B | Mus musculus (Mouse) | PR |
| P70587 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Rattus norvegicus (Rat) | PR |
| Q4V8G0 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Rattus norvegicus (Rat) | PR |
| Q80WD0 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Rattus norvegicus (Rat) | PR |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9SHI4 | RLP3 | Receptor-like protein 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5G5E0 | PIRL5 | Plant intracellular Ras-group-related LRR protein 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| B0JZ65 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q68F79 | lrrc8e | Volume-regulated anion channel subunit LRRC8E | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| B0R160 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MIPVTELRYF | ADTQPAYRIL | KPWWDVFTDY | ISIVMLMIAV | FGGTLQVTQD | KMICLPCKWV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TKDSCNDSFR | GWAASSPEPT | YPNSTVLPTP | DTGPTGIKYD | LDRHQYNYVD | AVCYENRLHW |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FAKYFPYLVL | LHTLIFLACS | NFWFKFPRTS | SKLEHFVSIL | LKCFDSPWTT | RALSETVVEE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SDPKPAFSKM | NGSMDKKSST | VSEDVEATVP | MLQRTKSRIE | QGIVDRSETG | VLDKKEGEQA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KALFEKVKKF | RTHVEEGDIV | YRLYMRQTII | KVIKFALIIC | YTVYYVHNIK | FDVDCTVDIE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SLTGYRTYRC | AHPLATLFKI | LASFYISLVI | FYGLICMYTL | WWMLRRSLKK | YSFESIREES |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SYSDIPDVKN | DFAFMLHLID | QYDPLYSKRF | AVFLSEVSEN | KLRQLNLNNE | WTLDKLRQRL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TKNAQDKLEL | HLFMLSGIPD | TVFDLVELEV | LKLELIPDVT | IPPSIAQLTG | LKELWLYHTA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| AKIEAPALAF | LRENLRALHI | KFTDIKEIPL | WIYSLKTLEE | LHLTGNLSAE | NNRYIVIDGL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RELKRLKVLR | LKSNLSKLPQ | VVTDVGVHLQ | KLSINNEGTK | LIVLNSLKKM | VNLTELELIR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| CDLERIPHSI | FSLHNLQEID | LKDNNLKTIE | EIISFQHLHR | LTCLKLWYNH | IAYIPIQIGN |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LTNLERLYLN | RNKIEKIPTQ | LFYCRKLRYL | DLSHNNLTFL | PADIGLLQNL | QNLAVTANRI |
| 730 | 740 | 750 | 760 | 770 | 780 |
| EALPPELFQC | RKLRALHLGN | NVLQSLPSRV | GELTNLTQIE | LRGNRLECLP | VELGECPLLK |
| 790 | 800 | ||||
| RSGLVVEEDL | FSTLPPEVKE | RLWRADKEQA |