Q80U19
Gene name |
Daam2 (Kiaa0381) |
Protein name |
Disheveled-associated activator of morphogenesis 2 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:76441 |
EC number |
|
Protein Class |
|
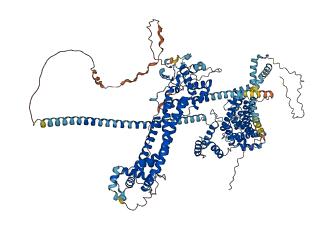
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
36-473 (DID domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Higashi T et al. (2010) "Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase", The Journal of biological chemistry, 285, 16231-8
Autoinhibited structure

Activated structure

1 structures for Q80U19
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q80U19-F1 | Predicted | AlphaFoldDB |
54 variants for Q80U19
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389382507 | 2 | A>T | No | EVA | |
| rs3389469092 | 14 | L>I | No | EVA | |
| rs3389470131 | 31 | H>Q | No | EVA | |
| rs3389460569 | 42 | I>N | No | EVA | |
| rs3389463917 | 68 | A>T | No | EVA | |
| rs3389469069 | 105 | R>C | No | EVA | |
| rs3389382585 | 113 | Q>P | No | EVA | |
| rs235945261 | 150 | D>E | No | EVA | |
| rs3389419251 | 187 | N>K | No | EVA | |
| rs3389382561 | 210 | T>I | No | EVA | |
| rs3389465008 | 215 | T>S | No | EVA | |
| rs3389465035 | 220 | L>M | No | EVA | |
| rs3389428527 | 220 | L>RDG* | No | EVA | |
| rs3389382580 | 222 | I>S | No | EVA | |
| rs3389470174 | 222 | I>V | No | EVA | |
| rs3389419257 | 225 | A>P | No | EVA | |
| rs246034793 | 245 | A>V | No | EVA | |
| rs3389428555 | 265 | G>D | No | EVA | |
| rs3405945528 | 306 | M>V | No | EVA | |
| rs3389460563 | 326 | K>Q | No | EVA | |
| rs3389469094 | 339 | D>N | No | EVA | |
| rs3389476460 | 343 | A>S | No | EVA | |
| rs3389463852 | 391 | N>Y | No | EVA | |
| rs3389382590 | 407 | Q>H | No | EVA | |
| rs3389470171 | 450 | K>R | No | EVA | |
| rs3389471163 | 477 | E>V | No | EVA | |
| rs3389471093 | 502 | G>E | No | EVA | |
| rs3389485803 | 505 | A>T | No | EVA | |
| rs3389463861 | 518 | P>S | No | EVA | |
| rs3389476426 | 522 | P>Q | No | EVA | |
| rs3389428533 | 525 | P>L | No | EVA | |
| rs3389485768 | 531 | L>M | No | EVA | |
| rs3389463887 | 541 | P>A | No | EVA | |
| rs3389471944 | 556 | P>L | No | EVA | |
| rs3389428561 | 616 | E>* | No | EVA | |
| rs3389469085 | 630 | Q>H | No | EVA | |
| rs3389471965 | 770 | E>D | No | EVA | |
| rs3389437252 | 779 | E>D | No | EVA | |
| rs3389460545 | 814 | K>* | No | EVA | |
| rs3389465002 | 852 | A>V | No | EVA | |
| rs3389437296 | 855 | N>K | No | EVA | |
| rs3389448612 | 892 | Y>* | No | EVA | |
| rs3389470114 | 894 | I>N | No | EVA | |
| rs3389437278 | 895 | M>K | No | EVA | |
| rs3389460557 | 945 | Y>F | No | EVA | |
| rs3389463859 | 980 | N>K | No | EVA | |
| rs3389428518 | 1004 | E>K | No | EVA | |
| rs3389448528 | 1036 | R>S | No | EVA | |
| rs3389481013 | 1038 | R>H | No | EVA | |
| rs47143619 | 1054 | R>T | No | EVA | |
| rs46545517 | 1056 | R>K | No | EVA | |
| rs46025208 | 1062 | G>S | No | EVA | |
| rs3389471991 | 1064 | L>R | No | EVA | |
| rs3389485746 | 1115 | Y>C | No | EVA |
No associated diseases with Q80U19
5 regional properties for Q80U19
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Formin, FH3 domain | 231 - 429 | IPR010472 |
| domain | Formin, GTPase-binding domain | 40 - 228 | IPR010473 |
| domain | Diaphanous autoregulatory (DAD) domain | 1065 - 1095 | IPR014767 |
| domain | Rho GTPase-binding/formin homology 3 (GBD/FH3) domain | 40 - 416 | IPR014768 |
| domain | Formin, FH2 domain | 595 - 1085 | IPR015425 |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin binding | Binding to monomeric or multimeric forms of actin, including actin filaments. |
| small GTPase binding | Binding to a small monomeric GTPase. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| determination of left/right symmetry | The establishment of an organism's body plan or part of an organism with respect to the left and right halves. The pattern can either be symmetric, such that the halves are mirror images, or asymmetric where the pattern deviates from this symmetry. |
| dorsal spinal cord development | The process whose specific outcome is the progression of the dorsal region of the spinal cord over time, from its formation to the mature structure. The dorsal region of the mature spinal cord contains neurons that process and relay sensory input. |
| negative regulation of oligodendrocyte differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of oligodendrocyte differentiation. |
| podocyte cell migration | The orderly movement of a podocyte from one site to another, often during the development of a multicellular organism or multicellular structure. A podocyte is a specialized kidney epithelial cell. |
| positive regulation of canonical Wnt signaling pathway | Any process that increases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| regulation of actin filament polymerization | Any process that modulates the frequency, rate or extent of the assembly of actin filaments by the addition of actin monomers to a filament. |
| regulation of canonical Wnt signaling pathway | Any process that modulates the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| regulation of filopodium assembly | Any process that modulates the frequency, rate or extent of the assembly of a filopodium, a thin, stiff protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal growth cone. |
| regulation of non-canonical Wnt signaling pathway | Any process that modulates the frequency, rate or extent of non-canonical Wnt signaling pathway. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0A1D5P556 | DAAM2 | Disheveled-associated activator of morphogenesis 2 | Gallus gallus (Chicken) | SS |
| O95466 | FMNL1 | Formin-like protein 1 | Homo sapiens (Human) | SS |
| Q27J81 | INF2 | Inverted formin-2 | Homo sapiens (Human) | EV |
| Q9Y4D1 | DAAM1 | Disheveled-associated activator of morphogenesis 1 | Homo sapiens (Human) | EV |
| Q86T65 | DAAM2 | Disheveled-associated activator of morphogenesis 2 | Homo sapiens (Human) | SS |
| Q0GNC1 | Inf2 | Inverted formin-2 | Mus musculus (Mouse) | SS |
| Q0QWG9 | Grid2ip | Delphilin | Mus musculus (Mouse) | PR |
| Q8BPM0 | Daam1 | Disheveled-associated activator of morphogenesis 1 | Mus musculus (Mouse) | PR |
| Q9C7S1 | FH12 | Formin-like protein 12 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LH02 | FH17 | Formin-like protein 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0C5K4 | FH21A | Putative formin-like protein 21a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FF14 | FH19 | Formin-like protein 19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MALRKRSPHG | LGFLCCFGGS | DLPEIDLRDS | HPLQYLEFSG | PIPNPEELNV | RFAELVDELD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LTDKNREAVF | ALPPEKKWQI | YCSKRKEQED | PNKLATSWPE | YYIDRINAMA | AMQNLYETED |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EETDKRNQVV | EDLKTALRTQ | PMRFVTRFID | LEGLTCLLNF | LRGMDHTTCE | SRIHTSLIGC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IKALMNNSQG | RAHVLAQPEA | ISIIAQSLRT | ENSKTKVAVL | EILGAVCLVP | GGHKKVLQAM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LHYQAYAAER | TRFQTLLNEL | DRSLGRYRDE | VNLKTAIMSF | INAVLNAGAG | EDNLEFRLHL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RYEFLMLGIQ | PVIDKLRQHE | NAILDKHLDF | FEMVRNEDDL | ELARRFDMVH | IDTKSASQMF |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ELIHKKLKHT | EAYPCLLSVL | HHCLQMPYKR | NGGYFQQWQL | LDRILQQIVL | QDERGVDPDL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| APLENFNVKN | IVNMLINENE | VKQWRDQAEK | FRKEHMELMS | RLERKERECE | TKTLEKEEMM |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RTLNKMKDKL | ARESQELRQA | RGQVAELVAR | HNESSTGPVS | SPPPPGGPLT | LSSSRTTNDL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PPPPPPLPFD | SCPPPPAPPL | PPGGPPIPPG | APPCFSSGPP | PSHDPFSSNE | APLRKKRIPQ |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PSHPLKSFNW | VKLNEERVSG | TVWNEIDDSQ | VFRILDLEDF | EKMFSAYQRH | QACMQEGPQR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| ERGNVRDGGA | ASRPLPAVEA | SAHRTEKASR | SMVSATGAKK | ELGSTEDIYI | TSRKVKELSV |
| 730 | 740 | 750 | 760 | 770 | 780 |
| IDGRRAQNCI | ILLSKLKLSN | DEIRQAILRM | DEQEDLAKDM | LEQLLKFIPE | KSDIDLLEEH |
| 790 | 800 | 810 | 820 | 830 | 840 |
| KHEIERMARA | DRFLYEMSRI | DHYQQRLQAL | FFKKKFQERL | AEAKPKVEAI | LLASRELTLS |
| 850 | 860 | 870 | 880 | 890 | 900 |
| QRLKQMLEVV | LAIGNFMNKG | QRGGAYGFRV | ASLNKIADTK | SSIDRNISLL | HYLIMILEKH |
| 910 | 920 | 930 | 940 | 950 | 960 |
| FPDILNMPSE | LKHLSEAAKV | NLAELEKEVS | ILRRGLRAVE | VELEYQRHQA | RDPNDKFVPV |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| MSDFITVSSF | SFSELEDQLN | EARDKFAKAL | THFGEQESKM | QPDEFFGIFD | TFLQAFLEAR |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| QDLEAMRRRK | EEDERRARME | FMLKEQREKE | RWQRQRKVLA | GGALEESGEF | DDLVSALRSG |
| 1090 | 1100 | 1110 | |||
| EVFDKDLSKF | KRNRKRPGSQ | VPEVTRERAI | NRLNY |