Q7Z4S6
Gene name |
KIF21A (KIAA1708, KIF2) |
Protein name |
Kinesin-like protein KIF21A |
Names |
Kinesin-like protein KIF2 , Renal carcinoma antigen NY-REN-62 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:55605 |
EC number |
|
Protein Class |
CHROMOSOME-ASSOCIATED KINESIN KIF4A-RELATED (PTHR47969) |
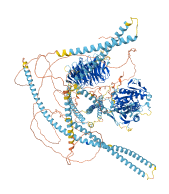
Descriptions
Kinesin (KIF) motor proteins play a major role in the transport of intracellular cargo and in the regulation of microtubule (MT) organization and dynamics in an ATP-dependent fashion. A prototypic kinesin includes a conserved globular N-terminal motor domain, which binds to MTs, hydrolyses ATP and converts its chemical energy to mechanical work. The kinesin motor domain is typically followed by a stalk domain often consisting of α-helical coiled-coil regions that are important for dimerization and a tail domain containing the binding sites for cargo or kinesin-regulatory proteins.
Intramolecular antiparallel coiled coil in the stalk domain autoinhibits the kinesin motor domain, preventing its interaction with microtubules (MTs) and ATP hydrolysis, thus regulating intracellular transport and MT dynamics. Interaction of the KIF21A regulatory domain with the KIF21B motor domain and sequence similarities to KIF7 and KIF27 strongly suggest a conservation of this regulatory mechanism in other kinesin-4 family members.
Autoinhibitory domains (AIDs)
Target domain |
7-379 (Kinesin motor domain) |
Relief mechanism |
Partner binding, Others |
Assay |
Structural analysis, Deletion assay, Split protein assay, Mutagenesis experiment |
Accessory elements
No accessory elements
References
Autoinhibited structure
Activated structure

1628 variants for Q7Z4S6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001110640 rs1227646139 |
66 | I>T | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
rs113878846 RCV002540804 RCV000910070 |
139 | A>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001110639 COSM3376233 rs141530856 |
190 | V>I | Congenital fibrosis of extraocular muscles type 1 pancreas [ClinVar, Cosmic] | Yes |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
COSM2065225 CA6511235 rs770064077 RCV000287713 COSM1361328 |
193 | R>H | Congenital fibrosis of extraocular muscles type 1 Variant assessed as Somatic; MODERATE impact. large_intestine [ClinVar, NCI-TCGA, Cosmic] | Yes |
ClinGen NCI-TCGA Cosmic cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV000389659 CA6511215 rs149328427 |
203 | Q>R | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV003311584 rs267603451 |
236 | Q>* | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
| VAR_074031 | 352 | D>E | CFEOM1; de novo mutation [UniProt] | Yes | UniProt |
|
RCV002535307 rs768580958 RCV000732988 |
356 | M>L | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs121912588 CA339982 RCV000002543 VAR_019399 |
356 | M>T | Congenital fibrosis of extraocular muscles type 1 CFEOM1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000319726 rs745692702 RCV000903904 CA6511100 |
387 | N>S | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000894701 CA6511073 RCV000262273 rs78616703 |
414 | G>R | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs375541289 RCV001113865 |
427 | M>V | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs886049345 CA10637334 RCV000354754 |
441 | A>T | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000302061 rs149219011 CA6510978 RCV000954280 |
543 | K>R | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA6510911 RCV000289132 RCV001701946 rs79089655 |
627 | G>V | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA6510910 rs765856120 RCV000399984 |
629 | S>I | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1555167299 RCV000497582 CA384401387 RCV003159607 |
664 | L>P | KIF21A-related condition [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs869025264 RCV000207402 CA351603 |
763 | V>M | Congenital aniridia [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs200645253 CA6510851 RCV000352193 |
769 | T>A | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000386473 rs779688115 CA6510787 |
811 | L>F | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs764601859 CA6510771 RCV000281670 |
831 | T>M | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs764601859 RCV001110553 COSM548312 |
831 | T>R | lung Congenital fibrosis of extraocular muscles type 1 [Cosmic, ClinVar] | Yes |
cosmic curated ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001110552 COSM4575362 COSM4575363 rs1458182449 |
835 | R>W | Congenital fibrosis of extraocular muscles type 1 Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
NCI-TCGA Cosmic ClinVar NCI-TCGA TOPMed dbSNP gnomAD |
|
rs145318959 COSM1163002 RCV001109765 |
887 | A>V | Congenital fibrosis of extraocular muscles type 1 pancreas [ClinVar, Cosmic] | Yes |
cosmic curated ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001109764 RCV000894861 rs149718388 |
895 | P>L | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs886049344 RCV000259437 CA10637329 |
915 | I>T | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs749644826 CA6510695 RCV000361240 RCV003362752 |
928 | R>C | Congenital fibrosis of extraocular muscles type 1 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000323422 rs886049343 CA10642076 |
933 | I>N | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
| VAR_074032 | 944 | E>Q | CFEOM1 [UniProt] | Yes | UniProt |
|
RCV000002546 rs267607200 CA115556 |
947 | M>I | Fibrosis of extraocular muscles, congenital, 3b [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000002545 VAR_019400 CA252285 rs121912590 |
947 | M>R | Congenital fibrosis of extraocular muscles type 1 CFEOM1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
| VAR_027021 | 947 | M>T | CFEOM1 [UniProt] | Yes | UniProt |
|
RCV000002544 rs121912589 VAR_019401 CA252284 |
947 | M>V | Congenital fibrosis of extraocular muscles type 1 CFEOM1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
| VAR_074033 | 954 | R>L | CFEOM1 [UniProt] | Yes | UniProt |
|
rs121912586 RCV002254900 RCV000002541 CA115555 VAR_019402 RCV000002540 |
954 | R>Q | Congenital fibrosis of extraocular muscles type 1 Fibrosis of extraocular muscles, congenital, 3b CFEOM1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000267056 rs121912585 RCV000002539 CA115554 RCV000002538 VAR_019403 RCV003398420 |
954 | R>W | Congenital fibrosis of extraocular muscles type 1 Variant assessed as Somatic; MODERATE impact. KIF21A-related condition Fibrosis of extraocular muscles, congenital, 3b CFEOM1 [ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl NCI-TCGA dbSNP |
| VAR_074034 | 1008 | A>P | CFEOM1 [UniProt] | Yes | UniProt |
|
VAR_019404 rs121912587 RCV000002542 CA252283 |
1010 | I>T | Congenital fibrosis of extraocular muscles type 1 CFEOM1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001109763 rs1946526630 |
1028 | V>I | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV000310515 RCV000882518 CA6510607 rs142268373 |
1050 | N>S | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs150294289 RCV000363848 CA6510558 RCV000904253 |
1111 | V>A | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001113783 rs1316892081 |
1185 | P>L | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV000406843 rs149075970 CA6510462 |
1214 | P>R | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000351593 rs75223821 CA6510438 |
1237 | E>D | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs747176706 RCV001113782 |
1281 | R>H | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000313138 CA10641128 rs886049342 |
1291 | T>N | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs1565724893 RCV000779102 |
1293 | S>missing | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1565724702 RCV000779101 |
1296 | N>missing | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs142879430 CA6510329 RCV000401978 |
1320 | R>K | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000281544 rs886049341 CA10642067 |
1422 | I>V | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
COSM1676830 COSM1676831 rs749290916 RCV002556197 RCV001112436 |
1423 | R>Q | Congenital fibrosis of extraocular muscles type 1 Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [ClinVar, NCI-TCGA] | Yes |
NCI-TCGA Cosmic ClinVar NCI-TCGA dbSNP gnomAD |
|
rs781214904 RCV000373719 CA6510226 |
1426 | T>M | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs748150530 CA6510167 RCV000335321 |
1491 | H>Y | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000893947 rs62000373 RCV001111982 |
1505 | S>R | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA10641118 RCV000287324 rs886049340 |
1510 | I>L | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA6510141 rs142038295 RCV000272319 |
1521 | M>R | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001111981 rs1942967439 |
1605 | G>E | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV000326200 CA10642059 rs886049339 |
1631 | A>T | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs373406994 RCV000268861 CA6510048 |
1654 | R>L | Congenital fibrosis of extraocular muscles type 1 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
| rs1939873512 | 2 | L>S | No | Ensembl | |
| rs998747931 | 2 | L>V | No |
TOPMed gnomAD |
|
| rs1939873512 | 2 | L>W | No | Ensembl | |
| rs2140543663 | 4 | A>S | No | Ensembl | |
| rs2140543663 | 4 | A>T | No | Ensembl | |
| rs901772088 | 4 | A>V | No |
TOPMed gnomAD |
|
| rs1269706987 | 5 | P>A | No | gnomAD | |
| rs942928510 | 7 | E>D | No |
TOPMed gnomAD |
|
| rs1283389822 | 9 | S>Y | No | gnomAD | |
| rs1040577254 | 10 | V>L | No |
TOPMed gnomAD |
|
| rs1040577254 | 10 | V>M | No |
TOPMed gnomAD |
|
| rs1383184252 | 14 | V>D | No | gnomAD | |
| rs1422024452 | 14 | V>I | No |
TOPMed gnomAD |
|
| rs1422024452 | 14 | V>L | No |
TOPMed gnomAD |
|
| rs753835660 | 16 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1949861277 | 19 | Q>* | No |
TOPMed gnomAD |
|
| rs1728873657 | 19 | Q>H | No | TOPMed | |
| rs1272892313 | 19 | Q>R | No |
TOPMed gnomAD |
|
| rs1196836806 | 20 | L>F | No | gnomAD | |
| TCGA novel | 20 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1344943070 | 21 | A>V | No | gnomAD | |
| rs1289975638 | 22 | K>E | No | gnomAD | |
| rs1036918539 | 23 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 25 | I>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766144623 | 27 | G>E | No |
ExAC gnomAD |
|
| rs864321718 | 28 | C>W | No | gnomAD | |
| rs146467179 | 29 | H>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs530120375 | 30 | I>L | No |
1000Genomes TOPMed gnomAD |
|
| rs751744115 | 30 | I>T | No |
ExAC gnomAD |
|
| rs530120375 | 30 | I>V | No |
1000Genomes TOPMed gnomAD |
|
| rs376379819 | 31 | C>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs775439890 | 34 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs775439890 | 34 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs200140335 | 36 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs200140335 | 36 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
|
rs964997318 COSM321238 |
38 | E>K | lung [Cosmic] | No |
cosmic curated TOPMed |
| rs752560589 | 39 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1338137842 | 40 | Q>H | No | TOPMed | |
| rs1161423002 | 41 | V>L | No | gnomAD | |
| rs777328454 | 42 | F>C | No |
ExAC gnomAD |
|
| rs747408878 | 44 | G>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 46 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201016471 | 47 | K>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1181921264 | 48 | A>P | No | gnomAD | |
| rs1181921264 | 48 | A>T | No | gnomAD | |
| rs778279184 | 50 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1949855738 | 54 | V>L | No | Ensembl | |
| rs749197486 | 56 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs780002236 | 57 | I>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 58 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM938955 COSM1651251 |
59 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1949854726 | 60 | Q>R | No | TOPMed | |
| rs1280148553 | 62 | E>G | No | gnomAD | |
| rs1196914694 | 64 | I>F | No |
TOPMed gnomAD |
|
| rs1566011738 | 64 | I>T | No | Ensembl | |
| rs1227646139 | 66 | I>S | No | gnomAD | |
| rs1949853648 | 67 | Q>R | No | TOPMed | |
| rs1949853098 | 69 | I>T | No | TOPMed | |
| rs764312133 | 69 | I>V | No |
ExAC gnomAD |
|
| rs375950963 | 70 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 74 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753023239 | 74 | E>K | No |
ExAC gnomAD |
|
| rs1949851973 | 75 | G>A | No | Ensembl | |
| TCGA novel | 76 | C>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1949851622 | 78 | E>G | No | TOPMed | |
| rs1411594155 | 81 | N>D | No |
TOPMed gnomAD |
|
| rs1026082131 | 81 | N>S | No | Ensembl | |
| rs1949850968 | 83 | T>A | No | Ensembl | |
| rs1398535026 | 85 | F>C | No | gnomAD | |
| rs1949850637 | 85 | F>L | No |
TOPMed gnomAD |
|
| TCGA novel | 87 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1476695099 | 98 | M>V | No | TOPMed | |
|
TCGA novel rs1949839293 |
99 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs757125466 | 100 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs757125466 | 100 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs752832786 | 101 | G>E | No |
ExAC gnomAD |
|
| rs1949838687 | 101 | G>R | No | TOPMed | |
| rs1949838112 | 104 | V>I | No | Ensembl | |
| rs755059417 | 106 | I>M | No |
ExAC gnomAD |
|
| rs1949836950 | 109 | E>D | No | TOPMed | |
| TCGA novel | 110 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766924557 | 112 | G>C | No |
ExAC gnomAD |
|
| rs1173428205 | 114 | I>V | No | TOPMed | |
| rs1949835705 | 116 | R>* | No | TOPMed | |
|
COSM1734426 rs750992981 |
116 | R>Q | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1340413916 | 120 | H>N | No |
TOPMed gnomAD |
|
| rs1949835141 | 120 | H>Q | No | TOPMed | |
| rs767747370 | 121 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs767747370 | 121 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs762274417 | 122 | F>Y | No |
ExAC gnomAD |
|
| rs1649896543 | 123 | K>E | No | TOPMed | |
| TCGA novel | 124 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772743219 | 124 | S>R | No |
TOPMed gnomAD |
|
| rs1949834077 | 126 | E>G | No | TOPMed | |
| rs773829948 | 126 | E>K | No |
ExAC gnomAD |
|
| rs768314730 | 127 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs774980465 | 127 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs774980465 | 127 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs574944321 | 128 | K>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs745772090 | 129 | K>I | No | ExAC | |
| rs781230981 | 129 | K>N | No |
ExAC TOPMed gnomAD |
|
|
COSM1639119 COSM1361329 |
129 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs148456295 | 130 | H>P | No |
ESP ExAC TOPMed gnomAD |
|
|
rs148456295 COSM1705389 |
130 | H>R | skin [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| TCGA novel | 130 | H>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1428463661 | 131 | I>L | No |
TOPMed gnomAD |
|
| rs746843180 | 131 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1428463661 | 131 | I>V | No |
TOPMed gnomAD |
|
| TCGA novel | 132 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM938954 COSM1651252 |
133 | I>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779218500 | 133 | I>V | No |
ExAC TOPMed gnomAD |
|
|
COSM4864123 COSM938953 |
134 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1442945887 | 135 | N>H | No |
TOPMed gnomAD |
|
| rs964671160 | 136 | G>E | No | Ensembl | |
| rs1209459274 | 138 | P>R | No | Ensembl | |
| rs113878846 | 139 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1249716902 | 140 | P>L | No | gnomAD | |
|
COSM4811402 COSM1299338 |
140 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756263422 | 141 | D>A | No |
ExAC gnomAD |
|
| rs750860679 | 142 | F>I | No |
ExAC gnomAD |
|
| rs1275019194 | 144 | V>A | No | gnomAD | |
| rs1189046583 | 144 | V>M | No |
TOPMed gnomAD |
|
| rs1380112604 | 146 | A>T | No | gnomAD | |
| rs1949828944 | 146 | A>V | No |
TOPMed gnomAD |
|
| rs928789507 | 151 | L>F | No | Ensembl | |
| rs1482050446 | 151 | L>H | No | gnomAD | |
| rs1949713428 | 152 | Y>C | No | TOPMed | |
| rs1949713249 | 154 | E>* | No | TOPMed | |
| rs2139132518 | 158 | D>A | No | 1000Genomes | |
|
COSM4869728 COSM938952 |
158 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV000896847 rs181888536 |
160 | F>L | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1158465554 | 161 | D>V | No | gnomAD | |
| rs1472980527 | 162 | T>I | No |
TOPMed gnomAD |
|
| rs1472980527 | 162 | T>N | No |
TOPMed gnomAD |
|
|
rs745978127 COSM4863659 COSM938951 |
164 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs781488871 | 164 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs781488871 | 164 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs757781644 | 166 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs757781644 | 166 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1949711468 | 168 | A>T | No | Ensembl | |
| rs752122331 | 169 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs764421420 | 170 | S>T | No |
ExAC gnomAD |
|
| rs761738616 | 171 | K>E | No | Ensembl | |
| rs1949710590 | 172 | K>I | No | Ensembl | |
| rs1949710247 | 172 | K>N | No | Ensembl | |
|
rs369178971 COSM1676845 |
173 | S>* | large_intestine [Cosmic] | No |
cosmic curated ESP gnomAD |
|
COSM4498370 COSM4498371 |
173 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1949710074 | 173 | S>P | No | TOPMed | |
| rs758804095 | 174 | N>I | No |
ExAC TOPMed |
|
| rs758804095 | 174 | N>S | No |
ExAC TOPMed |
|
| rs1565998337 | 178 | H>L | No | Ensembl | |
| rs758912579 | 180 | D>H | No |
ExAC gnomAD |
|
| rs758912579 | 180 | D>N | No |
ExAC gnomAD |
|
|
COSM4810790 COSM938950 |
180 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1741361 rs1949708537 |
182 | T>A | urinary_tract [Cosmic] | No |
cosmic curated Ensembl |
| rs776209935 | 182 | T>I | No |
ExAC gnomAD |
|
|
COSM3968182 rs776209935 COSM3968181 |
182 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC gnomAD |
|
COSM6072388 COSM6072389 |
184 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765869720 | 184 | G>R | No | ExAC | |
| rs1018223613 | 185 | I>V | No |
TOPMed gnomAD |
|
| rs1949707447 | 187 | T>I | No | TOPMed | |
| rs760646186 | 188 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs773328520 | 189 | G>D | No |
ExAC gnomAD |
|
| rs1949706418 | 191 | T>A | No | gnomAD | |
| rs945939403 | 191 | T>I | No |
TOPMed gnomAD |
|
| rs1949706052 | 192 | T>P | No | Ensembl | |
| rs775802404 | 193 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1439385947 | 198 | E>* | No | TOPMed | |
| rs1371062772 | 201 | M>L | No | gnomAD | |
| rs768395970 | 202 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs149328427 | 203 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs746150457 | 204 | C>F | No |
TOPMed gnomAD |
|
| rs746150457 | 204 | C>Y | No |
TOPMed gnomAD |
|
| rs148027037 | 207 | L>S | No |
1000Genomes ESP ExAC gnomAD |
|
| rs771004518 | 209 | A>D | No |
ExAC gnomAD |
|
| rs1949655132 | 211 | S>C | No | Ensembl | |
| rs1949655309 | 211 | S>T | No | Ensembl | |
| rs778195029 | 212 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1274485860 | 213 | T>K | No |
TOPMed gnomAD |
|
| rs1949654807 | 213 | T>P | No | Ensembl | |
| rs1592366272 | 214 | T>P | No | Ensembl | |
| rs891941469 | 216 | S>G | No |
TOPMed gnomAD |
|
| rs1949653684 | 218 | Q>H | No | Ensembl | |
| rs1949653327 | 221 | V>G | No | Ensembl | |
| rs1949653165 | 222 | Q>K | No | Ensembl | |
|
COSM1645567 COSM459526 rs567275114 |
225 | R>C | cervix Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1949652600 COSM84147 |
225 | R>H | pancreas [Cosmic] | No |
cosmic curated gnomAD |
| rs753495640 | 227 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1471828683 | 227 | H>Y | No | TOPMed | |
| TCGA novel | 228 | A>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM938949 rs779602428 COSM4867480 |
229 | I>V | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 231 | T>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1002148747 | 233 | H>N | No | TOPMed | |
| rs755633407 | 233 | H>R | No |
ExAC gnomAD |
|
| rs1336253126 | 234 | V>L | No | gnomAD | |
| rs1949650648 | 235 | C>Y | No | TOPMed | |
| rs968752395 | 236 | Q>H | No | TOPMed | |
| rs767206825 | 238 | R>T | No |
ExAC gnomAD |
|
| rs1398697802 | 239 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1949649363 | 240 | C>R | No | gnomAD | |
| COSM1322539 | 242 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1682477634 | 242 | Q>P | No | TOPMed | |
| rs935761437 | 243 | I>M | No | gnomAD | |
| rs1949648806 | 244 | D>A | No | gnomAD | |
| TCGA novel | 245 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1468192418 | 246 | D>E | No | gnomAD | |
| rs1164491796 | 246 | D>Y | No | gnomAD | |
| rs772197323 | 247 | N>S | No |
ExAC gnomAD |
|
| rs377116944 | 248 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs1396157941 | 249 | T>A | No | TOPMed | |
| TCGA novel | 250 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769022716 | 252 | K>R | No |
ExAC gnomAD |
|
|
rs749632095 RCV001355730 |
253 | I>T | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1949613554 | 254 | I>T | No | Ensembl | |
| rs1949612887 | 255 | S>A | No | gnomAD | |
| rs1949612542 | 258 | A>G | No | Ensembl | |
| rs1292335779 | 261 | N>S | No |
TOPMed gnomAD |
|
|
COSM4867251 COSM938948 |
262 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1949612006 | 264 | E>K | No |
TOPMed gnomAD |
|
| rs745576659 | 265 | T>N | No |
ExAC gnomAD |
|
| rs1949611433 | 267 | T>A | No | Ensembl | |
| rs1592362966 | 267 | T>N | No | Ensembl | |
|
COSM3688180 COSM3688181 |
269 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1592362890 | 270 | F>L | No | Ensembl | |
| rs1330960891 | 271 | H>R | No | gnomAD | |
|
COSM4858627 COSM693267 |
274 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1949609929 | 274 | D>N | No | gnomAD | |
| rs1272845957 | 276 | A>T | No | gnomAD | |
| rs138390624 | 278 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs937811368 | 283 | R>C | No |
1000Genomes TOPMed gnomAD |
|
|
COSM1676843 rs758156089 |
283 | R>H | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| TCGA novel | 283 | R>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1949608069 | 284 | T>I | No | Ensembl | |
| rs1949607102 | 289 | E>G | No | Ensembl | |
|
rs1017951046 COSM938947 COSM4868510 |
289 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs370066971 | 290 | R>K | No | ESP | |
| rs1478587281 | 291 | A>G | No | gnomAD | |
|
rs765082569 COSM1361327 COSM4786483 |
291 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| COSM1177175 | 293 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1592362425 | 294 | G>A | No | Ensembl | |
| rs750158577 | 294 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1177512217 | 295 | I>V | No | gnomAD | |
|
rs1949605140 COSM41532 |
296 | S>C | lung [Cosmic] | No |
cosmic curated TOPMed |
| rs1179670901 | 297 | I>V | No | TOPMed | |
| rs1949604804 | 298 | N>T | No | Ensembl | |
| rs773472902 | 299 | C>Y | No |
ExAC gnomAD |
|
| rs767553587 | 300 | G>E | No |
ExAC gnomAD |
|
|
COSM5643210 COSM5643209 |
300 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764295598 | 303 | A>P | No |
ExAC gnomAD |
|
| rs781240040 | 305 | G>D | No | Ensembl | |
| TCGA novel | 309 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs972411719 | 309 | S>R | No |
TOPMed gnomAD |
|
| rs1436855388 | 309 | S>R | No | gnomAD | |
| TCGA novel | 312 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1949361146 | 312 | G>R | No | Ensembl | |
| rs1949360938 | 313 | D>V | No | Ensembl | |
|
COSM693268 COSM4859377 |
315 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1351297429 | 318 | A>P | No |
TOPMed gnomAD |
|
| rs1344951736 | 318 | A>V | No | gnomAD | |
| rs1361472826 | 320 | H>R | No | gnomAD | |
| rs1204385293 | 321 | V>D | No | TOPMed | |
| rs1429561167 | 322 | P>A | No | gnomAD | |
| rs770246874 | 322 | P>L | No |
ExAC gnomAD |
|
| rs1490765240 | 323 | Y>C | No |
TOPMed gnomAD |
|
| rs1490765240 | 323 | Y>F | No |
TOPMed gnomAD |
|
| rs776365804 | 326 | S>T | No |
ExAC gnomAD |
|
| rs1949357660 | 329 | T>A | No |
TOPMed gnomAD |
|
| rs1024080688 | 329 | T>I | No | Ensembl | |
| rs1949357272 | 330 | R>G | No | gnomAD | |
| rs1833162130 | 332 | L>P | No |
TOPMed gnomAD |
|
| TCGA novel | 333 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1949356480 | 333 | Q>R | No | TOPMed | |
| TCGA novel | 334 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs971322383 | 337 | G>E | No |
TOPMed gnomAD |
|
| rs1183333668 | 337 | G>R | No |
TOPMed gnomAD |
|
| rs1454729970 | 338 | G>V | No | TOPMed | |
| TCGA novel | 338 | G>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1275123439 | 339 | N>S | No | gnomAD | |
| rs1949355192 | 340 | S>I | No | TOPMed | |
| rs772956810 | 343 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs771443337 | 343 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs772956810 | 343 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1948954674 | 344 | M>L | No | Ensembl | |
| rs1244589483 | 345 | I>M | No | TOPMed | |
| rs1948954301 | 345 | I>T | No | gnomAD | |
|
COSM187103 rs143166999 |
346 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1592319162 | 348 | V>D | No | Ensembl | |
| TCGA novel | 353 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948952455 | 356 | M>I | No | TOPMed | |
| rs768580958 | 356 | M>V | No |
ExAC TOPMed gnomAD |
|
|
rs992012618 COSM938945 COSM2065198 |
358 | T>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1948952236 | 358 | T>S | No | TOPMed | |
|
COSM6072390 COSM6072391 |
359 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779694955 | 361 | T>S | No |
ExAC gnomAD |
|
| TCGA novel | 363 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 363 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948950282 | 365 | A>S | No | TOPMed | |
| rs1592318930 | 365 | A>V | No | Ensembl | |
| rs778081595 | 366 | N>H | No |
ExAC TOPMed gnomAD |
|
| rs758353486 | 366 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs752828477 | 367 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs765481237 | 367 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs760108872 | 368 | A>T | No |
ExAC gnomAD |
|
| rs766620026 | 369 | R>G | No |
ExAC gnomAD |
|
| rs1948948515 | 369 | R>K | No | Ensembl | |
| rs761132468 | 371 | I>V | No |
ExAC gnomAD |
|
| rs767209233 | 372 | K>R | No |
ExAC gnomAD |
|
| rs1948947899 | 373 | N>S | No |
TOPMed gnomAD |
|
| rs761242353 | 375 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs761242353 | 375 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1948947313 | 376 | M>I | No | Ensembl | |
| rs1186045282 | 376 | M>T | No | gnomAD | |
| rs968830367 | 377 | V>A | No | gnomAD | |
| rs1948947149 | 377 | V>L | No | Ensembl | |
| rs1021616179 | 378 | N>S | No | gnomAD | |
| rs1948946798 | 378 | N>Y | No | gnomAD | |
| rs773844977 | 380 | D>N | No |
ExAC gnomAD |
|
| rs1302414390 | 381 | R>K | No | gnomAD | |
| rs1388678542 | 382 | A>T | No | gnomAD | |
| rs749195583 | 383 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs749195583 | 383 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1948945443 | 383 | S>I | No | Ensembl | |
| rs1349887483 | 385 | Q>E | No |
TOPMed gnomAD |
|
| rs369194542 | 385 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs769430367 | 386 | I>M | No |
ExAC gnomAD |
|
| TCGA novel | 386 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1019794380 | 387 | N>D | No | TOPMed | |
|
RCV001773300 rs2138873313 |
389 | L>I | No |
ClinVar Ensembl dbSNP |
|
|
COSM1676841 rs777791795 |
390 | R>C | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs758689865 | 390 | R>H | No |
ExAC TOPMed gnomAD |
|
|
RCV002223435 rs777791795 |
390 | R>S | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs748441314 | 391 | S>R | No |
ExAC gnomAD |
|
| rs1199323292 | 392 | E>K | No | gnomAD | |
|
COSM3811927 COSM3811926 |
394 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1287405376 | 395 | R>* | No | Ensembl | |
| rs779115969 | 395 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs267603450 | 396 | L>F | No |
ExAC gnomAD |
|
| rs1442245860 | 396 | L>R | No | gnomAD | |
| rs267603450 | 396 | L>V | No |
ExAC gnomAD |
|
| rs754258302 | 397 | Q>H | No |
ExAC gnomAD |
|
| rs76957342 | 397 | Q>P | No | Ensembl | |
| rs1464916547 | 398 | M>L | No | gnomAD | |
| rs1308921763 | 398 | M>R | No |
TOPMed gnomAD |
|
| rs1565945825 | 399 | E>K | No | gnomAD | |
| rs1948941362 | 401 | M>L | No |
TOPMed gnomAD |
|
| rs376944751 | 401 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948941362 | 401 | M>V | No |
TOPMed gnomAD |
|
| rs1278629407 | 402 | E>G | No | gnomAD | |
| rs931089309 | 404 | K>R | No | Ensembl | |
| rs898209512 | 405 | T>R | No | Ensembl | |
| rs1229330768 | 406 | G>S | No | gnomAD | |
| rs1948857862 | 408 | R>G | No | Ensembl | |
|
COSM4786877 COSM1322540 |
408 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs946615924 | 409 | I>V | No | Ensembl | |
| rs1948857484 | 410 | I>V | No | Ensembl | |
| rs1948857263 | 411 | D>A | No | Ensembl | |
| rs1244207399 | 412 | E>G | No | gnomAD | |
| rs750947190 | 412 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1948856710 | 413 | E>D | No | TOPMed | |
| rs78616703 | 414 | G>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs757651743 | 414 | G>D | No |
ExAC gnomAD |
|
| rs1416576490 | 415 | V>M | No |
TOPMed gnomAD |
|
| rs1948855733 | 416 | E>D | No |
TOPMed gnomAD |
|
| rs771773477 | 416 | E>G | No |
TOPMed gnomAD |
|
| rs1948855552 | 417 | S>G | No | TOPMed | |
| rs2138843354 | 418 | I>M | No | Ensembl | |
| rs879099697 | 418 | I>V | No | Ensembl | |
| rs751166979 | 419 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1948854860 | 420 | D>V | No | gnomAD | |
| rs1421287570 | 421 | M>I | No | gnomAD | |
|
RCV000910155 rs551748818 |
421 | M>V | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1160923543 | 422 | F>L | No | gnomAD | |
| rs1472208880 | 424 | E>D | No | gnomAD | |
| rs375541289 | 427 | M>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
TCGA novel rs1592312677 |
430 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| TCGA novel | 431 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 434 | N>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1471900499 COSM938944 COSM4870467 |
436 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs200436491 | 436 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs764712550 | 439 | I>V | No |
ExAC gnomAD |
|
| rs1948851642 | 441 | A>V | No | Ensembl | |
| rs1439743720 | 442 | M>I | No | gnomAD | |
| rs1948851469 | 442 | M>T | No | TOPMed | |
| rs368294102 | 443 | Q>* | No |
ESP TOPMed gnomAD |
|
| rs368294102 | 443 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
| rs144601000 | 444 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs144601000 | 444 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs2138841416 | 445 | T>A | No | Ensembl | |
| rs770807001 | 445 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs770807001 | 445 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1948850057 | 447 | D>G | No | Ensembl | |
| rs769003611 | 448 | A>V | No |
ExAC gnomAD |
|
| rs375768830 | 450 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs771402953 | 454 | T>A | No | Ensembl | |
| rs769811096 | 454 | T>I | No |
ExAC gnomAD |
|
| rs746352730 | 455 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs781683151 | 458 | S>I | No |
ExAC gnomAD |
|
| rs781683151 | 458 | S>N | No |
ExAC gnomAD |
|
| rs757563686 | 459 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1166484313 | 459 | D>G | No | gnomAD | |
| rs1183320157 | 461 | A>V | No | gnomAD | |
| rs1471332590 | 463 | H>N | No |
TOPMed gnomAD |
|
| rs1471332590 | 463 | H>Y | No |
TOPMed gnomAD |
|
| rs751929960 | 464 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs751929960 | 464 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1948846699 | 465 | L>I | No | gnomAD | |
| rs778178203 | 466 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1948846517 | 466 | A>T | No | Ensembl | |
|
COSM1511943 COSM6136709 COSM6136708 rs778178203 |
466 | A>V | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs758057629 | 467 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM5233394 COSM548310 |
467 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1333883973 | 469 | G>D | No |
TOPMed gnomAD |
|
| rs1333883973 | 469 | G>V | No |
TOPMed gnomAD |
|
| TCGA novel | 471 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778450822 | 471 | G>E | No |
ExAC gnomAD |
|
| rs754452058 | 473 | E>* | No |
ExAC gnomAD |
|
| rs1211798681 | 473 | E>G | No |
TOPMed gnomAD |
|
| rs1211798681 | 473 | E>V | No |
TOPMed gnomAD |
|
|
TCGA novel rs1948814189 |
474 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1486805804 | 476 | S>G | No | gnomAD | |
| rs1948813413 | 477 | N>S | No | gnomAD | |
| rs766053082 | 479 | I>T | No |
ExAC gnomAD |
|
| rs750432034 | 480 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs1948812656 | 481 | S>G | No | TOPMed | |
| TCGA novel | 481 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761541891 | 483 | I>V | No |
ExAC gnomAD |
|
|
COSM938943 COSM2065175 |
485 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs143300316 COSM108456 |
485 | E>K | skin [Cosmic] | No |
cosmic curated Ensembl |
| rs1948811700 | 485 | E>V | No | TOPMed | |
| rs916797963 | 486 | I>V | No |
TOPMed gnomAD |
|
|
COSM938942 rs1948811066 COSM4863679 |
487 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs1948810735 | 488 | D>A | No |
TOPMed gnomAD |
|
| rs1948810904 | 488 | D>Y | No | Ensembl | |
| rs2138827295 | 489 | L>V | No | Ensembl | |
| rs1179718800 | 491 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 493 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146440071 | 493 | L>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 493 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1453403699 | 495 | E>K | No |
TOPMed gnomAD |
|
| rs767618564 | 496 | S>N | No | Ensembl | |
| rs1948419475 | 497 | E>A | No | Ensembl | |
| rs773709604 | 498 | A>T | No |
ExAC gnomAD |
|
| rs1948418833 | 499 | V>G | No |
TOPMed gnomAD |
|
| rs772460803 | 500 | N>Y | No |
ExAC gnomAD |
|
|
COSM4467858 rs774562432 COSM4467857 |
504 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| TCGA novel | 506 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1481871548 | 507 | L>M | No | gnomAD | |
| rs1948417443 | 508 | T>A | No |
TOPMed gnomAD |
|
| rs1160277688 | 508 | T>R | No | TOPMed | |
| rs1948417225 | 510 | A>G | No | Ensembl | |
| rs1948416802 | 511 | T>A | No | Ensembl | |
| rs1452723005 | 512 | A>G | No |
TOPMed gnomAD |
|
| rs779882169 | 512 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs779882169 | 512 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs755633872 | 513 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1385293596 | 513 | R>K | No | gnomAD | |
| rs745377020 | 514 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs144047911 | 515 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs993302961 | 519 | G>E | No | Ensembl | |
| rs569736835 | 519 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1948413540 | 521 | S>P | No | TOPMed | |
| rs1592284814 | 522 | T>S | No | Ensembl | |
| rs1364104176 | 524 | S>F | No | gnomAD | |
| TCGA novel | 524 | S>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948412946 | 524 | S>T | No | Ensembl | |
| rs1349715518 | 525 | P>L | No | gnomAD | |
| rs758223938 | 525 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1431095676 | 527 | I>M | No |
TOPMed gnomAD |
|
| rs1948411879 | 527 | I>V | No | Ensembl | |
| rs960059923 | 529 | S>P | No | Ensembl | |
| rs1312032581 | 530 | S>* | No | TOPMed | |
| rs371442550 | 531 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948410647 | 532 | K>T | No | TOPMed | |
| rs867354889 | 533 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs139129865 | 534 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 535 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs923122059 | 535 | I>T | No | TOPMed | |
| rs2138685600 | 536 | E>K | No | Ensembl | |
| rs1245668808 | 537 | I>T | No | gnomAD | |
| rs1204999624 | 538 | I>K | No | gnomAD | |
| TCGA novel | 538 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760678283 | 539 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1358721057 | 539 | D>G | No |
TOPMed gnomAD |
|
|
rs1352108446 COSM275737 |
539 | D>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1231578129 | 540 | L>I | No | gnomAD | |
| rs1347396430 | 540 | L>P | No | gnomAD | |
| rs1347396430 | 540 | L>Q | No | gnomAD | |
| rs1363043598 | 541 | A>E | No | gnomAD | |
| rs1236806542 | 541 | A>P | No | gnomAD | |
| rs1236806542 | 541 | A>T | No | gnomAD | |
|
COSM2065161 COSM2065162 |
544 | D>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1381571381 | 544 | D>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs964475807 | 544 | D>V | No |
TOPMed gnomAD |
|
|
COSM938941 COSM4863628 |
544 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374696798 | 545 | L>V | No |
ESP ExAC gnomAD |
|
| rs1290382409 | 546 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
TCGA novel rs1134961 |
548 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| COSM261662 | 550 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774962499 | 550 | R>K | No |
ExAC gnomAD |
|
| rs968902446 | 551 | K>E | No | Ensembl | |
| rs769101427 | 552 | E>K | No |
ExAC gnomAD |
|
| rs1173833175 | 553 | K>N | No |
TOPMed gnomAD |
|
| rs1165249973 | 554 | R>K | No | gnomAD | |
| TCGA novel | 555 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1179328833 | 557 | K>R | No |
TOPMed gnomAD |
|
|
COSM2065160 COSM1361325 |
558 | R>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774683597 | 558 | R>M | No | Ensembl | |
| rs774683597 | 558 | R>T | No | Ensembl | |
| rs1443639940 | 560 | Q>H | No |
TOPMed gnomAD |
|
| rs1947904409 | 561 | K>Q | No | TOPMed | |
| TCGA novel | 562 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 563 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1947903943 | 564 | E>K | No | TOPMed | |
| rs1161243532 | 565 | S>N | No | TOPMed | |
| rs1024198839 | 567 | R>* | No |
TOPMed gnomAD |
|
| rs1258642320 | 567 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1482181587 | 568 | E>G | No | gnomAD | |
| rs191150586 | 569 | E>A | No |
1000Genomes TOPMed gnomAD |
|
| rs191150586 | 569 | E>G | No |
1000Genomes TOPMed gnomAD |
|
| rs988674826 | 570 | R>G | No |
TOPMed gnomAD |
|
| rs1254959041 | 571 | S>N | No | gnomAD | |
| rs1364807045 | 572 | V>A | No |
TOPMed gnomAD |
|
| rs775986011 | 573 | A>G | No |
ExAC gnomAD |
|
| rs770300950 | 576 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs148688136 | 576 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs148688136 | 576 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs746545050 | 577 | D>G | No |
ExAC TOPMed gnomAD |
|
|
COSM4870400 COSM938940 |
577 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1947492141 | 578 | N>S | No | Ensembl | |
| rs1325359832 | 578 | N>Y | No | TOPMed | |
| rs777619166 | 582 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs748137887 | 584 | E>D | No |
ExAC gnomAD |
|
| rs771990557 | 584 | E>G | No |
ExAC gnomAD |
|
| rs2138415603 | 585 | K>E | No | Ensembl | |
| rs778652648 | 585 | K>N | No |
ExAC gnomAD |
|
| rs1033430812 | 586 | K>E | No | TOPMed | |
| rs1399474327 | 589 | K>N | No |
TOPMed gnomAD |
|
|
COSM325630 rs1407646031 |
590 | G>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| rs754807741 | 590 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1407646031 | 590 | G>S | No |
TOPMed gnomAD |
|
| rs1947489076 | 592 | S>A | No | Ensembl | |
| rs147231078 | 592 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM938939 COSM4875650 |
593 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1468936451 | 595 | E>K | No | gnomAD | |
| rs906206403 | 597 | N>D | No |
TOPMed gnomAD |
|
| rs751656519 | 597 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1372629291 | 598 | E>A | No | gnomAD | |
| rs1592241041 | 599 | L>I | No | Ensembl | |
| TCGA novel | 600 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1947486606 | 600 | E>K | No | Ensembl | |
| TCGA novel | 601 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs373882115 | 601 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs758952493 | 601 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1173632775 | 604 | S>R | No | gnomAD | |
| rs1216438949 | 607 | V>L | No | gnomAD | |
| rs1448482009 | 608 | S>N | No | gnomAD | |
| rs1277934816 | 609 | D>G | No |
TOPMed gnomAD |
|
| rs1947450563 | 609 | D>H | No | Ensembl | |
| TCGA novel | 610 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1463663866 | 610 | H>R | No | TOPMed | |
| rs1241747341 | 610 | H>Y | No | TOPMed | |
| rs139879235 | 612 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1009678629 | 613 | E>A | No | gnomAD | |
| rs748099862 | 615 | E>Q | No | Ensembl | |
| TCGA novel | 617 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747284637 | 619 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs747284637 | 619 | E>Q | No |
ExAC TOPMed gnomAD |
|
|
rs1453778990 RCV001356104 |
621 | E>missing | No |
ClinVar dbSNP |
|
|
RCV000968687 rs750450259 |
621 | E>missing | No |
ClinVar dbSNP |
|
| rs1373965379 | 623 | D>E | No |
TOPMed gnomAD |
|
| rs1947446887 | 623 | D>G | No | gnomAD | |
| rs777970404 | 624 | I>T | No |
ExAC gnomAD |
|
| rs758583396 | 625 | D>G | No |
ExAC gnomAD |
|
| rs1168393894 | 626 | G>R | No |
TOPMed gnomAD |
|
| rs1427945712 | 626 | G>V | No | gnomAD | |
| COSM1322541 | 628 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1440893509 | 630 | S>P | No | TOPMed | |
| TCGA novel | 632 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1273575247 | 632 | E>K | No |
TOPMed gnomAD |
|
| rs2138399874 | 634 | D>E | No | Ensembl | |
| rs1353385135 | 634 | D>G | No |
TOPMed gnomAD |
|
| TCGA novel | 636 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2138399705 | 636 | E>D | No | Ensembl | |
| rs755232716 | 638 | D>G | No |
ExAC gnomAD |
|
| rs1947442598 | 639 | E>G | No | Ensembl | |
| rs138836723 | 642 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1373202204 | 643 | Y>F | No | TOPMed | |
| TCGA novel | 644 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1051668919 | 644 | Q>E | No | Ensembl | |
| rs1947399005 | 646 | D>E | No | TOPMed | |
| rs1337320262 | 646 | D>G | No | gnomAD | |
| rs2138386942 | 647 | L>V | No | 1000Genomes | |
| rs778929196 | 654 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs755495082 | 655 | A>T | No |
ExAC gnomAD |
|
|
COSM5223882 COSM1476515 |
657 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 659 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs897649790 | 660 | L>M | No | TOPMed | |
| rs897649790 | 660 | L>V | No | TOPMed | |
| rs1947397615 | 661 | I>V | No | Ensembl | |
| rs2138386375 | 662 | D>G | No | Ensembl | |
| rs2138386300 | 663 | E>Q | No | Ensembl | |
| rs1374420922 | 666 | N>K | No | gnomAD | |
| rs75160352 | 666 | N>T | No | Ensembl | |
|
COSM6136710 COSM6136711 |
668 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
TCGA novel rs754348492 |
672 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD NCI-TCGA |
| rs1696445047 | 676 | K>R | No | TOPMed | |
| rs750028358 | 677 | Q>H | No |
ExAC gnomAD |
|
| rs756499040 | 677 | Q>L | No |
ExAC gnomAD |
|
| rs1369964741 | 678 | Y>S | No | gnomAD | |
| rs1156747011 | 679 | E>V | No | gnomAD | |
|
COSM4863500 COSM938937 |
680 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs940074449 | 683 | M>L | No |
TOPMed gnomAD |
|
| rs2138384465 | 683 | M>T | No | Ensembl | |
| rs1180150511 | 684 | M>I | No |
TOPMed gnomAD |
|
| rs1408329611 | 684 | M>K | No | gnomAD | |
| rs1466720080 | 687 | H>R | No | gnomAD | |
|
COSM1511945 COSM6136713 rs751062866 COSM6136712 |
690 | R>L | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs751062866 | 690 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs761244403 | 690 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs201835314 | 691 | D>N | No | Ensembl | |
| rs1947391125 | 693 | Q>* | No | Ensembl | |
| rs1489790674 | 693 | Q>H | No | gnomAD | |
| rs1947390628 | 694 | L>I | No | Ensembl | |
| rs1394566563 | 697 | D>A | No |
TOPMed gnomAD |
|
| rs558091834 | 697 | D>H | No |
1000Genomes ExAC gnomAD |
|
| rs558091834 | 697 | D>Y | No |
1000Genomes ExAC gnomAD |
|
| rs762918844 | 698 | Q>L | No |
ExAC TOPMed gnomAD |
|
|
rs762918844 RCV000915150 |
698 | Q>R | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs192560751 | 699 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1309369202 | 700 | L>F | No | gnomAD | |
| rs1947389109 | 701 | Q>R | No | TOPMed | |
| rs1232370864 | 704 | G>S | No | gnomAD | |
|
COSM4776543 rs753650390 COSM305993 |
705 | S>L | Variant assessed as Somatic; MODERATE impact. endometrium haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1381584879 | 706 | V>I | No |
TOPMed gnomAD |
|
| rs1947339962 | 707 | E>K | No | Ensembl | |
|
COSM3460395 COSM3460396 |
708 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs560037174 | 709 | Y>H | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 712 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1299873101 | 712 | E>K | No | gnomAD | |
| rs1947339333 | 713 | K>E | No | gnomAD | |
| rs1374518123 | 714 | A>E | No | gnomAD | |
| TCGA novel | 714 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1432481022 | 714 | A>T | No | gnomAD | |
| rs1173694250 | 715 | K>T | No |
TOPMed gnomAD |
|
| rs756413250 | 716 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1467092894 | 716 | K>Q | No | gnomAD | |
|
COSM1361324 rs1193021160 COSM4747037 |
717 | V>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| TCGA novel | 717 | V>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750788580 | 718 | R>G | No |
ExAC gnomAD |
|
| rs781602588 | 718 | R>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 720 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1029964383 | 721 | Y>C | No |
TOPMed gnomAD |
|
| rs756882881 | 722 | E>G | No |
ExAC gnomAD |
|
| rs1592233191 | 722 | E>K | No | Ensembl | |
| rs751212958 | 724 | K>N | No |
ExAC gnomAD |
|
| rs1592233123 | 725 | L>P | No | Ensembl | |
|
COSM5923968 COSM5923967 |
726 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs80326094 | 726 | Q>K | No | Ensembl | |
| rs1947335931 | 726 | Q>R | No | Ensembl | |
| rs1281244118 | 727 | A>G | No | gnomAD | |
| rs1281244118 | 727 | A>V | No | gnomAD | |
| rs1202432059 | 728 | M>I | No | gnomAD | |
| rs1592233019 | 728 | M>R | No | TOPMed | |
| rs1592233019 | 728 | M>T | No | TOPMed | |
| COSM1322542 | 734 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM71339 | 735 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763620963 | 735 | L>I | No |
ExAC gnomAD |
|
| rs1947333914 | 736 | Q>H | No | TOPMed | |
| rs1226589623 | 736 | Q>K | No | gnomAD | |
| TCGA novel | 739 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1947333444 | 740 | K>Q | No | Ensembl | |
| TCGA novel | 740 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752626804 | 742 | H>D | No |
ExAC TOPMed gnomAD |
|
|
COSM693273 COSM4862114 |
742 | H>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752626804 | 742 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1486943329 | 743 | A>T | No |
TOPMed gnomAD |
|
| rs961965933 | 743 | A>V | No | Ensembl | |
| rs759615987 | 744 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs759615987 | 744 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs770896441 | 747 | K>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM1361322 COSM5096817 |
748 | N>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2138364197 | 748 | N>S | No | 1000Genomes | |
| rs1393671045 | 749 | Q>* | No |
TOPMed gnomAD |
|
| rs1393671045 | 749 | Q>K | No |
TOPMed gnomAD |
|
| rs1014776876 | 751 | Q>H | No | gnomAD | |
| rs774463961 | 752 | Y>* | No |
ExAC gnomAD |
|
| rs528921927 | 752 | Y>C | No |
ExAC gnomAD |
|
| rs528921927 | 752 | Y>F | No |
ExAC gnomAD |
|
| rs112367509 | 754 | K>R | No | Ensembl | |
| rs370078871 | 755 | Q>L | No |
ESP TOPMed gnomAD |
|
| rs775997715 | 757 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs1196503556 COSM4785921 COSM1361321 |
762 | D>G | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1362390539 | 762 | D>Y | No | TOPMed | |
| rs1354367889 | 763 | V>G | No | gnomAD | |
| rs1433614706 | 764 | M>I | No | Ensembl | |
| rs1318486272 | 765 | E>K | No | gnomAD | |
| rs1259861197 | 766 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1009420903 | 769 | T>I | No |
TOPMed gnomAD |
|
| TCGA novel | 769 | T>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4449441 rs1219032396 COSM4449440 |
769 | T>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs1947054603 | 771 | V>A | No | TOPMed | |
| rs1440816430 | 771 | V>I | No | gnomAD | |
|
COSM1676838 rs778271459 COSM938936 |
772 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs530439474 | 772 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1947053701 | 773 | L>R | No | Ensembl | |
|
COSM4812317 COSM1299337 |
777 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747887370 | 778 | K>R | No |
ExAC gnomAD |
|
|
COSM4845130 COSM4845129 rs778531745 |
780 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs754563722 | 781 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs754563722 | 781 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1947052317 | 782 | E>G | No | TOPMed | |
| rs766429919 | 783 | K>E | No |
ExAC gnomAD |
|
| rs1247162394 | 784 | A>D | No |
TOPMed gnomAD |
|
| rs1247162394 | 784 | A>G | No |
TOPMed gnomAD |
|
| rs1453700596 | 784 | A>S | No | gnomAD | |
| rs1947050886 | 785 | R>S | No | TOPMed | |
| rs755870837 | 786 | L>P | No |
ExAC gnomAD |
|
| rs1947050210 | 787 | T>A | No | gnomAD | |
| rs899183697 | 788 | E>A | No | gnomAD | |
| rs1251317083 | 789 | S>C | No | gnomAD | |
| rs750179951 | 792 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1947049086 | 793 | R>G | No | gnomAD | |
| rs373936264 | 796 | A>T | No |
ESP ExAC |
|
|
COSM1299336 COSM4810865 |
797 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1355495662 | 799 | K>R | No | gnomAD | |
| rs763062854 | 800 | K>N | No |
ExAC gnomAD |
|
|
COSM468306 COSM4857394 |
800 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 800 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1232994660 | 801 | D>N | No | gnomAD | |
| rs202062095 | 801 | D>V | No |
ExAC gnomAD |
|
| rs371582303 | 803 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1326193641 | 803 | R>H | No | gnomAD | |
| rs371582303 | 803 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs771426838 | 804 | K>E | No |
ExAC gnomAD |
|
| rs761306396 | 804 | K>R | No |
ExAC gnomAD |
|
| rs776751685 | 805 | R>K | No | Ensembl | |
| rs2138155038 | 807 | H>D | No | Ensembl | |
| rs778008456 | 807 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs749005274 | 809 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1946661491 | 810 | R>G | No | Ensembl | |
| rs1946661254 | 810 | R>K | No | TOPMed | |
| rs1946660385 | 813 | E>K | No | TOPMed | |
| rs769384831 | 814 | A>T | No |
ExAC gnomAD |
|
| rs887820875 | 815 | Q>P | No | TOPMed | |
|
COSM4931238 COSM4931237 |
817 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1416156635 | 819 | Q>E | No |
TOPMed gnomAD |
|
| rs1416156635 | 819 | Q>K | No |
TOPMed gnomAD |
|
|
COSM1676836 COSM1676837 |
820 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs745745130 | 821 | V>A | No | ExAC | |
| rs1285146797 | 822 | V>I | No |
TOPMed gnomAD |
|
| TCGA novel | 823 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781202644 | 824 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs781202644 | 824 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM4041549 rs558002876 COSM4041550 |
824 | R>H | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs751352937 | 825 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs751352937 | 825 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM1212403 rs777605261 |
825 | R>H | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs777605261 | 825 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs751352937 | 825 | R>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3671060 COSM3671061 |
826 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1272634893 | 829 | E>V | No | TOPMed | |
| rs1185704719 | 831 | T>A | No | TOPMed | |
| rs1946648663 | 832 | A>T | No | TOPMed | |
| rs1565819019 | 832 | A>V | No | Ensembl | |
| rs979633626 | 833 | L>F | No |
TOPMed gnomAD |
|
|
COSM693274 COSM4860732 rs979633626 |
833 | L>V | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs762662150 COSM2065130 COSM2065129 |
834 | R>C | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM415877 rs775230732 |
834 | R>H | urinary_tract [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
rs1351407142 COSM5840900 COSM5840899 |
835 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| TCGA novel | 836 | Q>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1946647257 | 837 | V>I | No | TOPMed | |
| rs866560111 | 838 | R>G | No | gnomAD | |
|
RCV000994895 rs1592198426 |
839 | P>L | No |
ClinVar Ensembl dbSNP |
|
| rs377418348 | 840 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1946646571 | 840 | M>V | No | TOPMed | |
| rs745390589 | 842 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs377100439 | 844 | V>G | No | Ensembl | |
| rs770845892 | 845 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs776073550 | 845 | A>S | No |
ExAC gnomAD |
|
| rs2138148644 | 846 | G>W | No | Ensembl | |
| rs78917178 | 847 | K>E | No |
ExAC gnomAD |
|
| rs756078781 | 848 | V>G | No | ExAC | |
| rs780333123 | 848 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1370378496 | 850 | R>Q | No |
TOPMed gnomAD |
|
| rs952754000 | 850 | R>W | No | gnomAD | |
| rs1300421030 | 852 | L>V | No | gnomAD | |
| rs750509204 | 853 | S>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 855 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767549345 | 855 | S>F | No |
ExAC gnomAD |
|
| rs757815392 | 856 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs764511984 | 857 | A>E | No |
ExAC gnomAD |
|
| TCGA novel | 857 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774938793 | 858 | P>L | No |
ExAC TOPMed |
|
| rs369381971 | 858 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs369381971 | 858 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1200130993 | 862 | T>A | No | gnomAD | |
| TCGA novel | 864 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 864 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1260086082 | 866 | A>V | No |
TOPMed gnomAD |
|
| rs373609055 | 870 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM4859702 rs746913383 COSM693275 |
873 | A>S | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 874 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1209001466 | 875 | R>G | No | gnomAD | |
| rs369621463 | 876 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 877 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1313920515 | 878 | A>V | No | gnomAD | |
| rs747977668 | 879 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs747977668 | 879 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1946636515 | 882 | M>I | No | Ensembl | |
| rs146428467 | 882 | M>V | No |
ESP ExAC gnomAD |
|
| rs1299585064 | 883 | R>G | No | gnomAD | |
| rs1401809989 | 883 | R>I | No |
TOPMed gnomAD |
|
| rs1401809989 | 883 | R>T | No |
TOPMed gnomAD |
|
| rs2138145260 | 884 | I>M | No | Ensembl | |
| rs781301465 | 888 | R>K | No |
ExAC gnomAD |
|
| rs781301465 | 888 | R>T | No |
ExAC gnomAD |
|
| rs757443106 | 889 | V>I | No |
ExAC TOPMed gnomAD |
|
|
COSM3986888 COSM3986889 |
890 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201913108 | 892 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1443528519 | 892 | L>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1946634018 | 894 | T>A | No | gnomAD | |
|
rs145533806 RCV000970245 |
894 | T>M | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs149718388 | 895 | P>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs377107935 | 896 | A>S | No |
ESP ExAC gnomAD |
|
| rs1946632527 | 897 | T>K | No | Ensembl | |
| rs1223175612 | 898 | N>D | No |
TOPMed gnomAD |
|
| rs1592197166 | 898 | N>T | No | Ensembl | |
| rs1946631660 | 900 | N>D | No |
TOPMed gnomAD |
|
| rs753468576 | 900 | N>K | No |
ExAC gnomAD |
|
|
COSM3460393 COSM3460394 |
901 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs755324647 | 902 | K>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 903 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1246547717 | 904 | Y>* | No |
TOPMed gnomAD |
|
| rs753476990 | 904 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1355412839 | 904 | Y>H | No | gnomAD | |
| rs982903798 | 907 | K>E | No |
TOPMed gnomAD |
|
| rs1307979882 | 908 | G>E | No |
TOPMed gnomAD |
|
| rs1307979882 | 908 | G>V | No |
TOPMed gnomAD |
|
| rs2138138634 | 910 | T>N | No | Ensembl | |
|
rs749918204 COSM1676834 |
912 | R>* | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs145303928 | 912 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs774185664 | 913 | V>E | No |
ExAC gnomAD |
|
| rs761776528 | 913 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1006504172 | 915 | I>M | No | TOPMed | |
| rs768448384 | 916 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs1179385674 | 916 | S>C | No | gnomAD | |
|
COSM3460392 COSM3460391 |
916 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768448384 | 916 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1196712876 | 919 | A>V | No | gnomAD | |
| rs201804365 | 920 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs747284586 COSM4041547 COSM4041548 |
920 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs747284586 | 920 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs747284586 | 920 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1351141435 | 922 | K>N | No | gnomAD | |
| rs1211233492 | 922 | K>T | No |
TOPMed gnomAD |
|
| rs777972416 | 924 | Q>H | No |
ExAC gnomAD |
|
| rs1284671239 | 924 | Q>R | No | gnomAD | |
|
COSM4041546 COSM4041545 |
925 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772077335 | 925 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs779305683 | 926 | L>F | No |
ExAC TOPMed |
|
| rs779305683 | 926 | L>I | No |
ExAC TOPMed |
|
| rs755299026 | 927 | E>* | No | ExAC | |
| rs755738527 | 928 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs755738527 | 928 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs750114694 | 929 | R>T | No |
ExAC gnomAD |
|
| rs1592195686 | 930 | V>G | No | Ensembl | |
| rs767011275 | 931 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1946610581 | 932 | D>N | No | Ensembl | |
| rs751455734 | 934 | I>V | No |
ExAC gnomAD |
|
| rs763933896 | 935 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1421552176 | 937 | K>N | No | gnomAD | |
| rs1946608931 | 942 | N>S | No |
TOPMed gnomAD |
|
| rs1409522933 | 943 | M>T | No |
TOPMed gnomAD |
|
| rs372545573 | 943 | M>V | No |
ESP TOPMed gnomAD |
|
| rs1056598799 | 945 | A>S | No |
TOPMed gnomAD |
|
| TCGA novel | 945 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 951 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376149427 | 952 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs745424578 | 952 | K>M | No | gnomAD | |
| rs773506249 | 953 | Q>K | No |
ExAC gnomAD |
|
| rs762149881 | 955 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs905295295 | 955 | E>G | No | TOPMed | |
| rs1279279983 | 956 | E>* | No | gnomAD | |
| rs1220693174 | 958 | T>I | No |
TOPMed gnomAD |
|
| rs1220693174 | 958 | T>K | No |
TOPMed gnomAD |
|
| rs774269490 | 960 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM3398689 COSM3398690 rs2138124848 |
960 | R>I | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
|
COSM3416808 rs142292357 COSM170145 |
961 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs142292357 | 961 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs199795220 | 961 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3416807 COSM3416806 |
962 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370441394 | 963 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM938935 COSM4868648 |
964 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770085648 | 964 | L>V | No |
ExAC gnomAD |
|
| rs1946577890 | 966 | K>E | No | TOPMed | |
| rs61733370 | 967 | R>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV000968252 rs61733370 |
967 | R>K | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs61733370 | 967 | R>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1397034944 | 969 | E>A | No |
TOPMed gnomAD |
|
| rs1439133785 | 969 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs144311378 | 971 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs746716611 | 971 | I>T | No |
ExAC gnomAD |
|
| rs144311378 | 971 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1180924393 | 972 | V>I | No | gnomAD | |
| rs777109447 | 973 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1946575414 | 974 | E>A | No | TOPMed | |
| rs1244501187 | 974 | E>D | No | gnomAD | |
| rs2138122956 | 975 | N>S | No | Ensembl | |
| rs1308444424 | 976 | G>E | No |
TOPMed gnomAD |
|
| rs1001479403 | 976 | G>R | No |
TOPMed gnomAD |
|
| COSM257236 | 978 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752627686 | 984 | N>D | No |
ExAC gnomAD |
|
| TCGA novel | 985 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs149602555 | 985 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs968203446 | 986 | N>S | No |
TOPMed gnomAD |
|
| rs1946573186 | 988 | E>A | No | TOPMed | |
| TCGA novel | 989 | M>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1312231703 | 989 | M>T | No | gnomAD | |
|
COSM6136714 COSM6136715 |
990 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 990 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1381400352 | 993 | T>S | No | gnomAD | |
| rs1946571907 | 994 | A>P | No | Ensembl | |
| rs377047702 | 996 | I>T | No |
ESP ExAC gnomAD |
|
| rs1331968401 | 997 | D>G | No | gnomAD | |
| rs61733764 | 997 | D>H | No | gnomAD | |
| rs61733764 | 997 | D>N | No | gnomAD | |
| rs761907530 | 999 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs369034538 | 1002 | S>G | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 1006 | C>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1946569843 | 1008 | A>D | No | gnomAD | |
| rs751774889 | 1009 | N>S | No | Ensembl | |
| rs751836346 | 1010 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed |
| rs764110171 | 1013 | M>T | No |
ExAC gnomAD |
|
| rs371835130 | 1014 | E>D | No | Ensembl | |
| rs1946568163 | 1015 | E>K | No |
TOPMed gnomAD |
|
| rs1245940570 | 1016 | A>T | No | TOPMed | |
| rs1305521189 | 1019 | E>G | No | gnomAD | |
| rs1192194635 | 1019 | E>K | No | gnomAD | |
| TCGA novel | 1020 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1490945887 | 1020 | G>D | No |
TOPMed gnomAD |
|
| rs145207490 | 1021 | E>K | No | 1000Genomes | |
| rs755957986 | 1022 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs146384175 | 1023 | L>S | No |
ESP TOPMed |
|
| rs113796726 | 1024 | D>E | No | gnomAD | |
| rs1946527284 | 1025 | V>A | No | TOPMed | |
| rs751673366 | 1028 | V>D | No |
ExAC gnomAD |
|
| rs1241096457 | 1029 | I>L | No |
TOPMed gnomAD |
|
| rs1241096457 | 1029 | I>V | No |
TOPMed gnomAD |
|
| rs1946525951 | 1031 | A>V | No | TOPMed | |
| rs1946525677 | 1032 | C>R | No | TOPMed | |
| rs763228459 | 1033 | T>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1034 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1946525030 | 1034 | L>V | No | Ensembl | |
| rs1946524402 | 1035 | T>K | No | Ensembl | |
| rs892495726 | 1035 | T>P | No | TOPMed | |
| rs1295910635 | 1037 | A>S | No | gnomAD | |
| rs1360570327 | 1038 | R>* | No |
TOPMed gnomAD |
|
| rs1946523260 | 1038 | R>Q | No |
TOPMed gnomAD |
|
| rs1946523030 | 1039 | Y>H | No | Ensembl | |
| rs771841179 | 1043 | H>Y | No |
ExAC gnomAD |
|
| rs1193181698 | 1047 | M>L | No |
TOPMed gnomAD |
|
| rs1946520573 | 1048 | G>D | No | gnomAD | |
| rs1488780871 | 1048 | G>R | No | gnomAD | |
| rs1263931298 | 1049 | I>V | No | gnomAD | |
| rs1311773456 | 1052 | G>D | No |
TOPMed gnomAD |
|
| rs761139342 | 1054 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs773694301 | 1055 | A>D | No |
ExAC gnomAD |
|
| rs773694301 | 1055 | A>G | No |
ExAC gnomAD |
|
| rs773694301 | 1055 | A>V | No |
ExAC gnomAD |
|
|
rs1230534151 COSM312336 |
1056 | A>S | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2138080683 | 1057 | Q>L | No | Ensembl | |
| rs761520066 | 1059 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1197714585 | 1059 | E>K | No | gnomAD | |
| rs1027109820 | 1061 | Q>* | No | Ensembl | |
| rs1339343134 | 1064 | V>G | No | gnomAD | |
| rs767995169 | 1065 | L>Q | No | ExAC | |
|
COSM1705381 rs915555359 |
1066 | E>K | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1239402355 | 1067 | G>V | No |
TOPMed gnomAD |
|
| rs748836406 | 1068 | R>* | No |
ExAC gnomAD |
|
| rs1375258659 | 1068 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1946446441 | 1070 | K>R | No | TOPMed | |
| TCGA novel | 1071 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774930561 | 1072 | T>A | No |
ExAC gnomAD |
|
| TCGA novel | 1073 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769757650 | 1073 | E>K | No |
ExAC gnomAD |
|
| rs745833102 | 1075 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1946445134 | 1078 | T>N | No | TOPMed | |
| rs780924477 | 1079 | Q>P | No |
ExAC gnomAD |
|
| rs780924477 | 1079 | Q>R | No |
ExAC gnomAD |
|
| rs1457631420 | 1080 | N>K | No | gnomAD | |
| rs1369122644 | 1081 | Q>L | No | gnomAD | |
| rs1437917259 | 1082 | L>I | No | gnomAD | |
| rs748309541 | 1083 | L>S | No |
ExAC gnomAD |
|
| rs992474024 | 1084 | F>L | No | TOPMed | |
| rs779110274 | 1084 | F>L | No |
ExAC gnomAD |
|
| rs1252460684 | 1084 | F>S | No | gnomAD | |
| rs1592185504 | 1085 | H>R | No | Ensembl | |
| rs1946442450 | 1085 | H>Y | No | TOPMed | |
| rs1946442071 | 1086 | M>L | No |
TOPMed gnomAD |
|
| rs960013288 | 1086 | M>T | No | Ensembl | |
| rs1946441676 | 1089 | E>D | No | TOPMed | |
| rs1946441471 | 1090 | K>E | No | Ensembl | |
| rs1946441247 | 1091 | A>T | No | Ensembl | |
| rs1946441045 | 1091 | A>V | No | TOPMed | |
| rs1451807193 | 1094 | N>S | No | TOPMed | |
| TCGA novel | 1095 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755218415 | 1097 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs753936272 | 1099 | A>T | No |
ExAC gnomAD |
|
| rs377193734 | 1101 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1408837029 | 1101 | L>P | No | TOPMed | |
| rs1408837029 | 1101 | L>Q | No | TOPMed | |
| rs377193734 | 1101 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM938934 COSM1651254 |
1102 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1946438829 | 1102 | G>D | No | gnomAD | |
| rs1423846263 | 1102 | G>S | No |
TOPMed gnomAD |
|
| rs1360492822 | 1104 | A>D | No |
TOPMed gnomAD |
|
|
COSM3416804 COSM3416803 |
1105 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1946438233 | 1106 | Q>R | No | TOPMed | |
| rs367901380 | 1109 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1294831190 | 1109 | D>G | No |
TOPMed gnomAD |
|
| rs1946396957 | 1109 | D>N | No | TOPMed | |
| rs1946396232 | 1110 | S>N | No | TOPMed | |
|
TCGA novel rs139033996 |
1110 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ESP ExAC TOPMed gnomAD |
| rs563027265 | 1111 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1167441024 | 1112 | P>L | No |
TOPMed gnomAD |
|
| rs1167441024 | 1112 | P>Q | No |
TOPMed gnomAD |
|
| rs781481134 | 1116 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs778149010 | 1117 | E>D | No |
ExAC TOPMed gnomAD |
|
|
rs140140022 RCV000912546 |
1117 | E>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs758701315 | 1119 | S>N | No |
ExAC gnomAD |
|
| rs2137914302 | 1121 | D>H | No | Ensembl | |
| rs201693220 | 1122 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs764790302 | 1123 | D>N | No |
ExAC gnomAD |
|
| rs1945902688 | 1123 | D>V | No | TOPMed | |
| rs796231505 | 1124 | A>T | No | TOPMed | |
| rs187723045 | 1125 | P>S | No |
1000Genomes TOPMed gnomAD |
|
| rs187723045 | 1125 | P>T | No |
1000Genomes TOPMed gnomAD |
|
| rs887878200 | 1127 | N>S | No |
TOPMed gnomAD |
|
| rs1945900859 | 1129 | P>L | No | Ensembl | |
| rs1945900594 | 1132 | E>D | No | gnomAD | |
| TCGA novel | 1133 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs965774800 | 1135 | T>M | No |
TOPMed gnomAD |
|
| rs753516869 | 1139 | D>H | No |
ExAC gnomAD |
|
| rs765831109 | 1140 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs1945847349 | 1141 | M>L | No | TOPMed | |
| rs952973809 | 1147 | V>A | No | Ensembl | |
| rs750311917 | 1147 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs761870399 | 1149 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1592156733 | 1150 | K>T | No | TOPMed | |
| rs372579199 | 1151 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3688179 COSM3688178 |
1152 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1179262275 | 1152 | K>R | No | gnomAD | |
| rs757111473 | 1153 | A>V | No |
ExAC gnomAD |
|
| rs1486236477 | 1154 | R>* | No |
TOPMed gnomAD |
|
|
rs986963607 COSM175284 |
1154 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1945431009 | 1155 | R>G | No | gnomAD | |
| TCGA novel | 1156 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs74778449 | 1156 | R>K | No | Ensembl | |
| rs1592139967 | 1157 | T>P | No | Ensembl | |
| rs763923109 | 1159 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs762845087 | 1161 | M>V | No |
ExAC gnomAD |
|
| rs1346942612 | 1165 | Y>C | No |
TOPMed gnomAD |
|
| rs761003662 | 1165 | Y>H | No |
ExAC TOPMed |
|
| rs1346942612 | 1165 | Y>S | No |
TOPMed gnomAD |
|
| rs1275921808 | 1167 | D>G | No | gnomAD | |
| rs1381122416 | 1169 | S>I | No | gnomAD | |
| rs1945427121 | 1169 | S>R | No | TOPMed | |
| rs1227919873 | 1171 | L>P | No | TOPMed | |
| rs1945426888 | 1171 | L>V | No | TOPMed | |
| rs772220692 | 1172 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs772220692 | 1172 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs762173645 | 1177 | T>A | No |
ExAC gnomAD |
|
| rs774856036 | 1177 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs554469530 | 1179 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs760073207 | 1179 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs575286430 | 1180 | A>G | No |
1000Genomes ExAC gnomAD |
|
| rs202078745 | 1180 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1180 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143083005 | 1181 | S>C | No |
ESP TOPMed gnomAD |
|
| rs143083005 | 1181 | S>F | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 1181 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs962843455 | 1182 | L>F | No |
TOPMed gnomAD |
|
| rs1188204192 | 1183 | P>L | No | gnomAD | |
|
COSM4041543 COSM4041544 |
1184 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1238651079 | 1186 | L>F | No |
TOPMed gnomAD |
|
|
COSM6072398 COSM6072399 |
1187 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945420673 | 1188 | P>L | No | TOPMed | |
| rs954946455 | 1188 | P>S | No | TOPMed | |
| rs954946455 | 1188 | P>T | No | TOPMed | |
| rs1262449822 | 1189 | V>I | No |
TOPMed gnomAD |
|
| rs1262449822 | 1189 | V>L | No |
TOPMed gnomAD |
|
| rs1945419876 | 1190 | A>G | No | Ensembl | |
| rs1203990826 | 1190 | A>T | No | gnomAD | |
| rs1945419614 | 1192 | G>R | No | Ensembl | |
| rs1945419112 | 1193 | Q>* | No |
TOPMed gnomAD |
|
|
COSM4833152 COSM4833151 |
1194 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1390414058 | 1194 | E>V | No | TOPMed | |
|
COSM4832278 COSM4832279 |
1195 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945418877 | 1195 | I>V | No | TOPMed | |
| rs1945418640 | 1199 | T>P | No | TOPMed | |
| rs1451001972 | 1200 | E>G | No | TOPMed | |
| rs1945417891 | 1201 | T>A | No | Ensembl | |
| rs1249207861 | 1202 | S>I | No |
TOPMed gnomAD |
|
| rs1026567466 | 1203 | G>D | No | Ensembl | |
| TCGA novel | 1203 | G>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1203 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1015969607 | 1204 | T>I | No |
TOPMed gnomAD |
|
| rs756814252 | 1204 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs756814252 | 1204 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1394783161 | 1205 | S>T | No |
TOPMed gnomAD |
|
| rs1945415757 | 1208 | E>G | No | Ensembl | |
| rs557018562 | 1209 | K>E | No |
1000Genomes TOPMed gnomAD |
|
| TCGA novel | 1209 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758466226 | 1210 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1945414686 | 1211 | L>F | No | TOPMed | |
| rs1945414445 | 1212 | S>F | No | TOPMed | |
| rs866768006 | 1213 | P>L | No | TOPMed | |
| rs1945413905 | 1213 | P>S | No | TOPMed | |
| rs149075970 | 1214 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs149075970 | 1214 | P>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1945412683 | 1216 | G>D | No |
TOPMed gnomAD |
|
| rs571247823 | 1218 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1219 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs147620197 COSM3398687 |
1220 | K>E | central_nervous_system [Cosmic] | No |
cosmic curated ESP TOPMed gnomAD |
| rs552970913 | 1220 | K>R | No |
1000Genomes TOPMed gnomAD |
|
|
COSM1476513 COSM4813862 |
1221 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1222 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1592138934 | 1222 | G>S | No | Ensembl | |
| rs1945411222 | 1222 | G>V | No | Ensembl | |
| rs1945411013 | 1223 | S>R | No | Ensembl | |
| rs2137771966 | 1224 | I>S | No | Ensembl | |
| rs1945030197 | 1225 | S>C | No | Ensembl | |
| rs1945029662 | 1227 | Q>K | No | Ensembl | |
| rs1438184694 | 1227 | Q>R | No | gnomAD | |
| rs2137660591 | 1230 | L>P | No | Ensembl | |
| rs780767170 | 1230 | L>V | No | Ensembl | |
| rs1252175888 | 1231 | S>L | No | gnomAD | |
| rs1219822918 | 1233 | K>E | No | gnomAD | |
| rs750548981 | 1234 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs750548981 | 1234 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs1291959900 | 1234 | K>T | No | gnomAD | |
|
COSM1361319 COSM5091775 rs761074592 |
1235 | I>F | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs767779686 | 1235 | I>M | No |
ExAC gnomAD |
|
| rs145259176 | 1236 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs746483961 | 1237 | E>Q | No | Ensembl | |
| rs1334390547 | 1239 | S>A | No | TOPMed | |
| rs927536292 | 1239 | S>F | No |
TOPMed gnomAD |
|
| TCGA novel | 1239 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs147861618 | 1240 | P>L | No |
ESP gnomAD |
|
| rs763411761 | 1240 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs980738403 | 1241 | V>L | No | TOPMed | |
| rs776087480 | 1242 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1399041505 | 1242 | T>I | No | gnomAD | |
| rs776087480 | 1242 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs776087480 | 1242 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs569992788 | 1245 | K>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs759940671 | 1245 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs375504040 | 1246 | A>P | No |
ESP TOPMed gnomAD |
|
| rs375504040 | 1246 | A>T | No |
ESP TOPMed gnomAD |
|
| rs1592122713 | 1246 | A>V | No | Ensembl | |
| rs202059694 | 1248 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs202059694 | 1248 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs746695893 | 1249 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs1479940083 | 1252 | K>I | No | gnomAD | |
| rs1269171184 | 1254 | K>R | No |
TOPMed gnomAD |
|
| rs772806269 | 1257 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1432997726 | 1261 | S>A | No | gnomAD | |
| rs1367190751 | 1262 | D>G | No | gnomAD | |
| rs2137588687 | 1262 | D>N | No | Ensembl | |
| rs1334651917 | 1263 | S>P | No |
TOPMed gnomAD |
|
| rs1944784548 | 1264 | G>E | No | gnomAD | |
| rs1944784548 | 1264 | G>V | No | gnomAD | |
| rs201205072 | 1265 | T>A | No | TOPMed | |
| rs1592111055 | 1266 | S>L | No | Ensembl | |
| rs1944782965 | 1268 | A>G | No | Ensembl | |
| rs1177753954 | 1268 | A>S | No |
TOPMed gnomAD |
|
| rs773795201 | 1269 | S>N | No |
ExAC gnomAD |
|
| rs1592110965 | 1271 | S>L | No | Ensembl | |
| rs768618302 | 1272 | P>R | No |
ExAC gnomAD |
|
| rs1944781508 | 1273 | P>S | No | TOPMed | |
| COSM116656 | 1274 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1183539058 | 1275 | S>P | No | gnomAD | |
| rs1756239087 | 1276 | P>S | No | TOPMed | |
|
rs139695269 COSM1476512 |
1279 | R>Q | breast [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs1384710859 | 1280 | P>H | No | gnomAD | |
| rs1384710859 | 1280 | P>R | No | gnomAD | |
|
COSM4948628 COSM1361318 rs769601725 |
1281 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs752800426 | 1282 | N>S | No |
ExAC gnomAD |
|
| rs1245207332 | 1284 | L>P | No |
TOPMed gnomAD |
|
| rs1245207332 | 1284 | L>R | No |
TOPMed gnomAD |
|
|
rs150630573 RCV000902855 |
1285 | N>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1944775541 | 1286 | V>G | No |
TOPMed gnomAD |
|
| TCGA novel | 1287 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755468804 | 1289 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs887165821 COSM4545797 COSM4545798 |
1289 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs755468804 | 1289 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1944774159 | 1290 | L>F | No | Ensembl | |
| rs1944773853 | 1290 | L>P | No | TOPMed | |
| TCGA novel | 1291 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs886049342 | 1291 | T>I | No | gnomAD | |
| rs766790592 | 1292 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs371178638 | 1294 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs371178638 | 1294 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1565724787 | 1295 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs767268089 | 1297 | T>A | No |
ExAC gnomAD |
|
|
COSM3811921 COSM415878 |
1298 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1944769629 | 1299 | V>I | No |
TOPMed gnomAD |
|
| rs768249226 | 1300 | Q>H | No |
ExAC gnomAD |
|
| rs1944768695 | 1301 | Q>K | No |
TOPMed gnomAD |
|
| rs1944768441 | 1303 | K>E | No | TOPMed | |
| rs1276329048 | 1303 | K>M | No | TOPMed | |
| rs11171717 | 1304 | S>C | No |
ExAC gnomAD |
|
| rs11171717 | 1304 | S>Y | No |
ExAC gnomAD |
|
| rs763602499 | 1305 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs1012793782 | 1306 | E>G | No | Ensembl | |
| rs2137515461 | 1307 | S>N | No | Ensembl | |
| rs201618273 | 1310 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA |
| rs2137515299 | 1310 | S>P | No | Ensembl | |
| rs1944493117 | 1311 | L>V | No | Ensembl | |
| rs769695817 | 1312 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs531164985 | 1313 | E>D | No |
ExAC TOPMed gnomAD |
|
|
COSM3986886 COSM3986887 |
1315 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM2065060 rs781410152 |
1317 | S>F | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1485764024 COSM3460390 |
1318 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1459792944 | 1318 | S>P | No |
TOPMed gnomAD |
|
| rs1459792944 | 1318 | S>T | No |
TOPMed gnomAD |
|
| rs142879430 | 1320 | R>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1460980013 | 1321 | G>V | No | TOPMed | |
| rs1944033331 | 1323 | I>M | No | Ensembl | |
|
RCV003237995 rs2137384157 |
1323 | I>T | No |
ClinVar Ensembl dbSNP |
|
| rs1944033582 | 1323 | I>V | No |
TOPMed gnomAD |
|
| rs1444013812 | 1324 | N>S | No | gnomAD | |
| rs1410743282 | 1325 | P>A | No | gnomAD | |
| rs1944032519 | 1325 | P>L | No | TOPMed | |
| rs1944032279 | 1326 | F>Y | No | TOPMed | |
| rs1375008050 | 1328 | A>T | No |
TOPMed gnomAD |
|
| rs778528102 | 1329 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs762576003 | 1330 | K>E | No |
TOPMed gnomAD |
|
| rs1424107928 | 1332 | I>M | No | gnomAD | |
| rs1944030545 | 1333 | R>K | No | TOPMed | |
| rs753262148 | 1333 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1565688929 | 1334 | A>V | No | Ensembl | |
| rs931031780 | 1337 | L>F | No | gnomAD | |
|
rs931031780 RCV001758147 |
1337 | L>I | No |
ClinVar dbSNP gnomAD |
|
| rs1180639291 | 1338 | Q>H | No | gnomAD | |
| rs1472747319 | 1339 | C>G | No | gnomAD | |
| rs1426376893 | 1339 | C>W | No | Ensembl | |
| rs755967049 | 1340 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1471226955 | 1340 | I>V | No | Ensembl | |
| rs767162841 | 1342 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs761705954 | 1343 | A>V | No |
ExAC gnomAD |
|
| rs1256275079 | 1344 | E>K | No |
TOPMed gnomAD |
|
| rs751467047 | 1346 | H>L | No |
ExAC gnomAD |
|
| rs1348979571 | 1347 | T>A | No |
TOPMed gnomAD |
|
| rs930559700 | 1348 | K>R | No |
TOPMed gnomAD |
|
|
COSM938931 COSM4874936 |
1349 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776530688 | 1350 | V>A | No | ExAC | |
| rs1352492048 | 1350 | V>M | No | gnomAD | |
| rs1036294156 | 1352 | C>S | No |
TOPMed gnomAD |
|
| rs1036294156 | 1352 | C>Y | No |
TOPMed gnomAD |
|
| rs771063766 | 1353 | V>L | No |
ExAC gnomAD |
|
| rs1225263157 | 1354 | D>N | No |
TOPMed gnomAD |
|
| rs1225263157 | 1354 | D>Y | No |
TOPMed gnomAD |
|
| rs1261194298 | 1355 | S>A | No |
TOPMed gnomAD |
|
| rs1261194298 | 1355 | S>P | No |
TOPMed gnomAD |
|
| rs980462939 | 1356 | T>I | No |
TOPMed gnomAD |
|
| rs761028766 | 1357 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs139664756 | 1358 | D>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM6072400 COSM6072401 |
1358 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139664756 | 1358 | D>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs748495397 | 1359 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1445476814 | 1359 | L>P | No |
TOPMed gnomAD |
|
| rs773322171 | 1360 | L>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs980893693 | 1361 | F>L | No |
TOPMed gnomAD |
|
| rs1413652783 | 1362 | T>S | No |
TOPMed gnomAD |
|
| TCGA novel | 1364 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1479807856 | 1365 | K>Q | No | gnomAD | |
|
COSM2065044 rs756337041 COSM2065043 |
1367 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs562228123 | 1367 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756337041 | 1367 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1300102262 | 1370 | K>E | No | TOPMed | |
| rs1943836099 | 1371 | V>L | No | TOPMed | |
| rs774856058 | 1372 | W>* | No |
ExAC gnomAD |
|
| rs762262699 | 1372 | W>R | No |
ExAC TOPMed gnomAD |
|
|
COSM4874236 COSM938930 |
1375 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2137327940 | 1377 | G>V | No | Ensembl | |
| TCGA novel | 1378 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1367079034 | 1378 | Q>R | No | gnomAD | |
| rs1565677895 | 1379 | E>K | No | Ensembl | |
| rs1943833125 | 1380 | I>R | No | Ensembl | |
| rs543953019 | 1381 | M>I | No |
1000Genomes TOPMed |
|
| rs916696180 | 1381 | M>L | No |
TOPMed gnomAD |
|
| rs1943832504 | 1381 | M>T | No | TOPMed | |
| rs916696180 | 1381 | M>V | No |
TOPMed gnomAD |
|
| rs764380324 | 1383 | L>R | No |
ExAC gnomAD |
|
| rs1416916210 | 1383 | L>V | No | gnomAD | |
| rs775777214 | 1384 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs775777214 | 1384 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs1407687334 | 1385 | G>R | No |
TOPMed gnomAD |
|
| rs1407687334 | 1385 | G>S | No |
TOPMed gnomAD |
|
| rs769466593 | 1385 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1487519274 | 1386 | H>Y | No | TOPMed | |
| rs1943828624 | 1387 | P>L | No | Ensembl | |
| rs2137326315 | 1388 | N>H | No | Ensembl | |
| rs2137326172 | 1389 | N>H | No | Ensembl | |
| rs745549119 | 1389 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs2137326008 | 1389 | N>K | No | Ensembl | |
| rs745549119 | 1389 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1943827345 | 1391 | V>G | No | Ensembl | |
| rs139013210 | 1391 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM75151 rs746875582 COSM4947701 |
1396 | C>Y | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1202745618 | 1397 | N>D | No | gnomAD | |
| rs925281361 | 1398 | Y>C | No |
TOPMed gnomAD |
|
| rs925281361 | 1398 | Y>F | No |
TOPMed gnomAD |
|
| rs771051669 | 1399 | T>I | No | Ensembl | |
| TCGA novel | 1399 | T>Y | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2137325012 | 1401 | L>W | No | Ensembl | |
| rs758321044 | 1403 | F>L | No |
ExAC gnomAD |
|
| rs1344148767 | 1404 | T>S | No | gnomAD | |
| rs2137324478 | 1405 | V>G | No | Ensembl | |
| rs747949177 | 1405 | V>I | No |
ExAC TOPMed gnomAD |
|
|
COSM1299335 COSM4812061 |
1406 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2137324413 | 1406 | S>L | No | Ensembl | |
| rs778610172 | 1407 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs2137324202 | 1408 | S>F | No | Ensembl | |
| rs1379688467 | 1409 | Y>S | No |
TOPMed gnomAD |
|
| rs2137323943 | 1410 | I>M | No | Ensembl | |
| rs190902651 | 1410 | I>V | No |
1000Genomes ExAC gnomAD |
|
| rs750669566 | 1412 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs2137323697 | 1413 | W>* | No | Ensembl | |
| rs1943821797 | 1413 | W>G | No | TOPMed | |
| rs2137323534 | 1418 | S>L | No | Ensembl | |
| rs201680308 | 1418 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs974476147 | 1419 | A>T | No | gnomAD | |
| rs757338165 | 1420 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs757338165 | 1420 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs2137323180 | 1421 | C>G | No | Ensembl | |
| rs751709193 | 1421 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs751709193 | 1421 | C>Y | No |
ExAC TOPMed gnomAD |
|
|
rs1164718381 COSM1705378 COSM1705377 |
1423 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs764649339 | 1424 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1291824309 | 1425 | L>V | No |
TOPMed gnomAD |
|
| rs781392170 | 1428 | S>L | No |
ExAC gnomAD |
|
| rs1297147858 | 1429 | G>A | No |
TOPMed gnomAD |
|
| rs1297147858 | 1429 | G>D | No |
TOPMed gnomAD |
|
| rs777756778 | 1430 | Q>E | No |
ExAC gnomAD |
|
| rs758899586 | 1430 | Q>H | No |
ExAC gnomAD |
|
| rs1943610952 | 1433 | L>F | No | TOPMed | |
| TCGA novel | 1434 | G>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1943610184 | 1435 | D>E | No | Ensembl | |
| rs1305397666 | 1435 | D>N | No |
TOPMed gnomAD |
|
| rs1226311136 | 1435 | D>V | No | TOPMed | |
| rs144392523 | 1436 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs144392523 | 1436 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs200589960 | 1437 | C>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs200589960 | 1437 | C>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs200589960 | 1437 | C>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1943608853 | 1439 | A>P | No | TOPMed | |
| rs1943608853 | 1439 | A>T | No | TOPMed | |
| rs766103846 | 1440 | S>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1440 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs185075080 | 1440 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs766103846 | 1440 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs772735255 | 1441 | T>I | No |
ExAC gnomAD |
|
|
COSM4947791 rs771502610 COSM1361315 |
1443 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs761755690 COSM3416801 COSM3416802 |
1443 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1943606674 | 1444 | T>I | No | gnomAD | |
| rs1259827334 | 1445 | V>A | No |
TOPMed gnomAD |
|
| TCGA novel | 1446 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1565665239 | 1447 | I>T | No | Ensembl | |
| rs774163102 | 1448 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs2137272949 | 1450 | G>E | No | Ensembl | |
| rs369666213 | 1452 | N>D | No |
ESP ExAC gnomAD |
|
| rs369666213 | 1452 | N>H | No |
ESP ExAC gnomAD |
|
| TCGA novel | 1454 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749073888 | 1454 | I>V | No |
ExAC gnomAD |
|
| rs1427117718 | 1455 | N>D | No |
TOPMed gnomAD |
|
| rs771111885 | 1458 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs747264491 | 1458 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs548227296 | 1459 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1943602400 | 1460 | N>H | No | Ensembl | |
| rs530018336 | 1460 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1255084857 | 1461 | P>S | No | gnomAD | |
|
RCV000894967 rs748963082 |
1463 | G>D | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs568874833 | 1464 | T>I | No |
1000Genomes ExAC gnomAD |
|
| rs1943600335 | 1465 | F>V | No | TOPMed | |
| rs1943599861 | 1466 | L>F | No | TOPMed | |
| rs1366357542 | 1466 | L>P | No | gnomAD | |
| rs1300196237 | 1467 | Y>C | No |
TOPMed gnomAD |
|
| rs1300196237 | 1467 | Y>F | No |
TOPMed gnomAD |
|
| rs1290102511 | 1467 | Y>H | No |
TOPMed gnomAD |
|
| rs1007813259 | 1470 | S>F | No | Ensembl | |
| rs1381233597 | 1471 | G>E | No |
TOPMed gnomAD |
|
| rs149234238 | 1474 | V>I | No |
ESP TOPMed gnomAD |
|
| rs149234238 | 1474 | V>L | No |
ESP TOPMed gnomAD |
|
|
COSM6072403 COSM6072402 |
1475 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1467627382 | 1476 | M>I | No | gnomAD | |
| rs1372844076 | 1478 | D>N | No | TOPMed | |
| TCGA novel | 1479 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755587423 | 1479 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1187806057 | 1484 | S>C | No | gnomAD | |
| TCGA novel | 1484 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1425121304 | 1485 | T>A | No | gnomAD | |
|
COSM3460387 COSM3460386 |
1486 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1332172227 | 1490 | G>E | No | TOPMed | |
| TCGA novel | 1493 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755501569 | 1493 | G>S | No |
ExAC gnomAD |
|
| rs1364901418 | 1494 | P>H | No | TOPMed | |
| rs756554787 | 1494 | P>S | No |
ExAC gnomAD |
|
| rs1222473844 | 1496 | M>I | No | TOPMed | |
| rs750868436 | 1496 | M>L | No |
ExAC gnomAD |
|
| rs750868436 | 1496 | M>V | No |
ExAC gnomAD |
|
| rs767064636 | 1498 | L>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1499 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751006090 | 1500 | V>A | No |
ExAC gnomAD |
|
| TCGA novel | 1502 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs559104078 | 1503 | I>N | No |
1000Genomes ExAC gnomAD |
|
| rs559104078 | 1503 | I>S | No |
1000Genomes ExAC gnomAD |
|
| rs931221280 | 1503 | I>V | No | Ensembl | |
| rs1352452589 | 1505 | S>G | No |
TOPMed gnomAD |
|
| rs2137197164 | 1505 | S>N | No | Ensembl | |
| rs775422512 | 1506 | G>E | No |
ExAC gnomAD |
|
| rs1943301002 | 1506 | G>R | No | Ensembl | |
| rs375548931 | 1508 | D>N | No |
ESP ExAC gnomAD |
|
| rs1565649083 | 1508 | D>V | No | Ensembl | |
| rs375548931 | 1508 | D>Y | No |
ESP ExAC gnomAD |
|
| rs1158024107 | 1515 | K>E | No | gnomAD | |
| rs1023072936 | 1516 | D>G | No |
TOPMed gnomAD |
|
| rs1023072936 | 1516 | D>V | No |
TOPMed gnomAD |
|
| rs1472614748 | 1517 | H>R | No | gnomAD | |
| TCGA novel | 1518 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1518 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1378693683 COSM368030 |
1521 | M>I | lung [Cosmic] | No |
cosmic curated gnomAD |
| rs756823322 | 1521 | M>V | No |
ExAC gnomAD |
|
| rs1023408067 | 1524 | V>I | No | Ensembl | |
| rs1324792116 | 1525 | T>A | No | TOPMed | |
| rs1943123168 | 1525 | T>I | No | TOPMed | |
|
COSM4899635 COSM4899636 |
1526 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777209231 | 1528 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs777209231 | 1528 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1448038974 | 1529 | L>P | No | gnomAD | |
| rs1209759462 | 1530 | G>A | No | gnomAD | |
| rs1943120717 | 1533 | S>R | No | Ensembl | |
| rs1362336549 | 1536 | H>Y | No | gnomAD | |
| rs1262925987 | 1537 | N>S | No |
TOPMed gnomAD |
|
| rs759553232 | 1540 | P>A | No |
ExAC gnomAD |
|
| rs1943119388 | 1540 | P>L | No | Ensembl | |
|
COSM3871521 COSM3871522 |
1541 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1592028243 | 1541 | P>S | No | Ensembl | |
| rs753536074 | 1543 | Y>C | No |
ExAC gnomAD |
|
| rs1943118349 | 1544 | D>G | No | Ensembl | |
| rs766218025 | 1546 | I>V | No |
ExAC gnomAD |
|
| rs780066416 | 1547 | E>D | No |
ExAC gnomAD |
|
| TCGA novel | 1548 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1346548419 | 1548 | A>V | No | gnomAD | |
| rs774716605 | 1551 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs1413375205 | 1551 | I>V | No |
TOPMed gnomAD |
|
| rs763113489 | 1553 | G>V | No |
ExAC gnomAD |
|
| rs769068732 | 1553 | G>W | No |
ExAC gnomAD |
|
| rs775761627 | 1554 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs775761627 | 1554 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1429547646 | 1557 | F>C | No | TOPMed | |
| rs1235926181 | 1558 | S>I | No | gnomAD | |
| rs1943113825 | 1559 | G>R | No | TOPMed | |
| rs1478111510 | 1560 | S>A | No | gnomAD | |
| rs982138535 | 1560 | S>F | No | TOPMed | |
| rs2137153523 | 1563 | N>T | No | 1000Genomes | |
| rs746422413 | 1565 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs771343710 | 1569 | D>G | No |
ExAC gnomAD |
|
| rs746600657 | 1572 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1381721611 | 1572 | Q>P | No | TOPMed | |
| rs191093660 | 1574 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1273856513 | 1574 | D>G | No | gnomAD | |
| TCGA novel | 1574 | D>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1734419 COSM1734420 |
1577 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1157266802 | 1578 | Q>* | No | gnomAD | |
| rs748814688 | 1579 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs780029810 | 1580 | P>S | No |
ExAC gnomAD |
|
| rs756170780 | 1581 | N>D | No |
ExAC gnomAD |
|
| rs750213364 | 1582 | A>S | No |
ExAC gnomAD |
|
| rs1942972642 | 1583 | H>Y | No |
TOPMed gnomAD |
|
| rs1490739730 | 1584 | K>R | No | TOPMed | |
| rs1189183372 | 1585 | D>G | No |
TOPMed gnomAD |
|
| rs1189183372 | 1585 | D>V | No |
TOPMed gnomAD |
|
| rs767452046 | 1586 | W>L | No |
ExAC gnomAD |
|
| rs758649013 | 1587 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs765166685 | 1591 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1942969389 | 1592 | V>L | No | gnomAD | |
| rs2137122371 | 1593 | V>L | No | Ensembl | |
| rs141385146 | 1596 | H>Y | No |
ESP ExAC gnomAD |
|
| rs537501726 | 1598 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs537501726 | 1598 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1565631039 | 1600 | L>P | No | gnomAD | |
| rs945173948 | 1600 | L>V | No | Ensembl | |
| rs1439161364 | 1603 | C>G | No | Ensembl | |
|
COSM4041535 COSM4041536 |
1606 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766876071 | 1606 | G>D | No |
ExAC gnomAD |
|
| rs1437592707 | 1615 | T>A | No | gnomAD | |
| rs773513818 | 1615 | T>I | No |
ExAC gnomAD |
|
| rs773513818 | 1615 | T>N | No |
ExAC gnomAD |
|
| rs893540223 | 1616 | F>S | No | TOPMed | |
| rs1347330994 | 1617 | M>T | No | gnomAD | |
| rs1942965110 | 1621 | E>D | No | TOPMed | |
| rs772438071 | 1623 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs747732382 | 1624 | G>C | No |
ExAC gnomAD |
|
| rs10243 | 1624 | G>D | No | TOPMed | |
| rs747732382 | 1624 | G>S | No |
ExAC gnomAD |
|
| rs1161324215 | 1625 | H>N | No |
TOPMed gnomAD |
|
| rs943051896 | 1625 | H>R | No | Ensembl | |
| rs768248912 | 1626 | D>G | No |
ExAC gnomAD |
|
| rs1942962671 | 1627 | S>T | No | gnomAD | |
| rs1454548070 | 1629 | I>F | No |
TOPMed gnomAD |
|
| rs1942961799 | 1629 | I>N | No | TOPMed | |
| rs1454548070 | 1629 | I>V | No |
TOPMed gnomAD |
|
| rs748809944 | 1630 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1307686317 | 1631 | A>D | No |
TOPMed gnomAD |
|
| rs148144546 | 1632 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756075691 | 1632 | I>M | No |
ExAC gnomAD |
|
| rs148144546 | 1632 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4862248 COSM693277 |
1634 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781080527 | 1636 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs781080527 | 1636 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs1301181207 | 1638 | H>D | No |
TOPMed gnomAD |
|
| rs1942957889 | 1641 | T>S | No | Ensembl | |
| rs1373843147 | 1642 | A>P | No | gnomAD | |
| rs1373843147 | 1642 | A>S | No | gnomAD | |
| rs1942105749 | 1644 | D>E | No | Ensembl | |
| rs1048446665 | 1645 | D>E | No | TOPMed | |
| rs777458254 | 1646 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs777458254 | 1646 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM220207 rs755174363 |
1646 | R>Q | NS Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs559949605 | 1647 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs559949605 | 1647 | T>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1240016204 | 1648 | V>E | No |
TOPMed gnomAD |
|
| rs780041023 | 1648 | V>M | No |
ExAC gnomAD |
|
| rs993800051 | 1649 | R>I | No |
TOPMed gnomAD |
|
| rs1348946004 | 1651 | W>S | No |
TOPMed gnomAD |
|
| rs896750929 | 1652 | K>R | No |
TOPMed gnomAD |
|
| rs1942102415 | 1653 | A>G | No | Ensembl | |
| rs376346007 | 1654 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
rs373406994 COSM4152803 |
1654 | R>H | kidney [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs376346007 | 1654 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs370691140 | 1655 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1942101240 | 1655 | N>Y | No | Ensembl | |
| rs199566972 | 1656 | L>F | No | Ensembl | |
| rs1340484297 | 1656 | L>S | No | gnomAD | |
|
COSM370736 rs1253389910 |
1657 | Q>P | lung [Cosmic] | No |
cosmic curated gnomAD |
| TCGA novel | 1658 | D>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1243077166 | 1659 | G>S | No | gnomAD | |
| rs1266144841 | 1660 | Q>H | No | Ensembl | |
| rs181255946 | 1660 | Q>L | No | 1000Genomes | |
| TCGA novel | 1661 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1321308319 | 1661 | I>T | No |
TOPMed gnomAD |
|
| rs1235189460 | 1662 | S>P | No |
TOPMed gnomAD |
|
| rs893438891 | 1665 | G>E | No | Ensembl | |
| rs769657363 | 1666 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs769657363 | 1666 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs769657363 | 1666 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs775014213 | 1667 | L>M | No |
ExAC gnomAD |
|
| rs769639690 | 1668 | G>E | No |
ExAC gnomAD |
|
| rs769639690 | 1668 | G>V | No |
ExAC gnomAD |
|
| rs770949743 | 1671 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs770949743 | 1671 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs770949743 | 1671 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs777563049 | 1672 | A>V | No |
ExAC gnomAD |
|
| rs771768221 | 1673 | S>N | No |
ExAC gnomAD |
|
| rs1409209542 | 1674 | N>D | No | gnomAD | |
|
COSM4934137 COSM4934136 |
1674 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780239746 | 1675 | N>L | No |
ExAC TOPMed gnomAD |
1 associated diseases with Q7Z4S6
[MIM: 135700]: Fibrosis of extraocular muscles, congenital, 1 (CFEOM1)
A congenital ocular motility disorder marked by restrictive ophthalmoplegia affecting extraocular muscles innervated by the oculomotor and/or trochlear nerves. It is clinically characterized by anchoring of the eyes in downward gaze, ptosis, and backward tilt of the head. Patients affected by congenital fibrosis of extraocular muscles type 1 show an absence of the superior division of the oculomotor nerve (cranial nerve III) and corresponding oculomotor subnuclei. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A congenital ocular motility disorder marked by restrictive ophthalmoplegia affecting extraocular muscles innervated by the oculomotor and/or trochlear nerves. It is clinically characterized by anchoring of the eyes in downward gaze, ptosis, and backward tilt of the head. Patients affected by congenital fibrosis of extraocular muscles type 1 show an absence of the superior division of the oculomotor nerve (cranial nerve III) and corresponding oculomotor subnuclei. . Note=The disease is caused by variants affecting the gene represented in this entry.
8 regional properties for Q7Z4S6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | WD40 repeat | 1336 - 1414 | IPR001680-1 |
| repeat | WD40 repeat | 1440 - 1478 | IPR001680-2 |
| repeat | WD40 repeat | 1481 - 1523 | IPR001680-3 |
| repeat | WD40 repeat | 1531 - 1615 | IPR001680-4 |
| repeat | WD40 repeat | 1615 - 1655 | IPR001680-5 |
| domain | Kinesin motor domain | 7 - 379 | IPR001752 |
| conserved_site | WD40 repeat, conserved site | 1360 - 1374 | IPR019775 |
| conserved_site | Kinesin motor domain, conserved site | 268 - 279 | IPR019821 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47969 | CHROMOSOME-ASSOCIATED KINESIN KIF4A-RELATED |
| PANTHER Subfamily | PTHR47969:SF30 | KINESIN MOTOR DOMAIN-CONTAINING PROTEIN |
| PANTHER Protein Class |
cytoskeletal protein
microtubule binding motor protein microtubule or microtubule-binding cytoskeletal protein |
|
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| kinesin complex | Any complex that includes a dimer of molecules from the kinesin superfamily, a group of related proteins that contain an extended region of predicted alpha-helical coiled coil in the main chain that likely produces dimerization. The native complexes of several kinesin family members have also been shown to contain additional peptides, often designated light chains as all of the noncatalytic subunits that are currently known are smaller than the chain that contains the motor unit. Kinesin complexes generally possess a force-generating enzymatic activity, or motor, which converts the free energy of the gamma phosphate bond of ATP into mechanical work. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ankyrin repeat binding | Binding to an ankyrin repeat of a protein. Ankyrin repeats are tandemly repeated modules of about 33 amino acids; each repeat folds into a helix-loop-helix structure with a beta-hairpin/loop region projecting out from the helices at a 90-degree angle, and repeats stack to form an L-shaped structure. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| microtubule motor activity | A motor activity that generates movement along a microtubule, driven by ATP hydrolysis. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| microtubule-based movement | A microtubule-based process that results in the movement of organelles, other microtubules, or other cellular components. Examples include motor-driven movement along microtubules and movement driven by polymerization or depolymerization of microtubules. |
20 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O95239 | KIF4A | Chromosome-associated kinesin KIF4A | Homo sapiens (Human) | SS |
| Q2VIQ3 | KIF4B | Chromosome-associated kinesin KIF4B | Homo sapiens (Human) | SS |
| P33176 | KIF5B | Kinesin-1 heavy chain | Homo sapiens (Human) | EV |
| O60282 | KIF5C | Kinesin heavy chain isoform 5C | Homo sapiens (Human) | EV |
| Q12840 | KIF5A | Kinesin heavy chain isoform 5A | Homo sapiens (Human) | EV |
| Q2M1P5 | KIF7 | Kinesin-like protein KIF7 | Homo sapiens (Human) | EV |
| Q86VH2 | KIF27 | Kinesin-like protein KIF27 | Homo sapiens (Human) | SS |
| O75037 | KIF21B | Kinesin-like protein KIF21B | Homo sapiens (Human) | EV |
| Q9P2E2 | KIF17 | Kinesin-like protein KIF17 | Homo sapiens (Human) | EV |
| O00139 | KIF2A | Kinesin-like protein KIF2A | Homo sapiens (Human) | PR |
| Q9NQT8 | KIF13B | Kinesin-like protein KIF13B | Homo sapiens (Human) | EV |
| Q9H1H9 | KIF13A | Kinesin-like protein KIF13A | Homo sapiens (Human) | SS |
| O43896 | KIF1C | Kinesin-like protein KIF1C | Homo sapiens (Human) | SS |
| Q7M6Z4 | Kif27 | Kinesin-like protein KIF27 | Mus musculus (Mouse) | SS |
| Q9QXL1 | Kif21b | Kinesin-like protein KIF21B | Mus musculus (Mouse) | SS |
| B7ZNG0 | Kif7 | Kinesin-like protein KIF7 | Mus musculus (Mouse) | SS |
| Q9QXL2 | Kif21a | Kinesin-like protein KIF21A | Mus musculus (Mouse) | EV SS |
| Q7M6Z5 | Kif27 | Kinesin-like protein KIF27 | Rattus norvegicus (Rat) | SS |
| F1M5N7 | Kif21b | Kinesin-like protein KIF21B | Rattus norvegicus (Rat) | SS |
| Q58G59 | kif7 | Kinesin-like protein kif7 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLGAPDESSV | RVAVRIRPQL | AKEKIEGCHI | CTSVTPGEPQ | VFLGKDKAFT | FDYVFDIDSQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QEQIYIQCIE | KLIEGCFEGY | NATVFAYGQT | GAGKTYTMGT | GFDVNIVEEE | LGIISRAVKH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LFKSIEEKKH | IAIKNGLPAP | DFKVNAQFLE | LYNEEVLDLF | DTTRDIDAKS | KKSNIRIHED |
| 190 | 200 | 210 | 220 | 230 | 240 |
| STGGIYTVGV | TTRTVNTESE | MMQCLKLGAL | SRTTASTQMN | VQSSRSHAIF | TIHVCQTRVC |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PQIDADNATD | NKIISESAQM | NEFETLTAKF | HFVDLAGSER | LKRTGATGER | AKEGISINCG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LLALGNVISA | LGDKSKRATH | VPYRDSKLTR | LLQDSLGGNS | QTIMIACVSP | SDRDFMETLN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TLKYANRARN | IKNKVMVNQD | RASQQINALR | SEITRLQMEL | MEYKTGKRII | DEEGVESIND |
| 430 | 440 | 450 | 460 | 470 | 480 |
| MFHENAMLQT | ENNNLRVRIK | AMQETVDALR | SRITQLVSDQ | ANHVLARAGE | GNEEISNMIH |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SYIKEIEDLR | AKLLESEAVN | ENLRKNLTRA | TARAPYFSGS | STFSPTILSS | DKETIEIIDL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AKKDLEKLKR | KEKRKKKRLQ | KLEESNREER | SVAGKEDNTD | TDQEKKEEKG | VSERENNELE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| VEESQEVSDH | EDEEEEEEEE | EDDIDGGESS | DESDSESDEK | ANYQADLANI | TCEIAIKQKL |
| 670 | 680 | 690 | 700 | 710 | 720 |
| IDELENSQKR | LQTLKKQYEE | KLMMLQHKIR | DTQLERDQVL | QNLGSVESYS | EEKAKKVRSE |
| 730 | 740 | 750 | 760 | 770 | 780 |
| YEKKLQAMNK | ELQRLQAAQK | EHARLLKNQS | QYEKQLKKLQ | QDVMEMKKTK | VRLMKQMKEE |
| 790 | 800 | 810 | 820 | 830 | 840 |
| QEKARLTESR | RNREIAQLKK | DQRKRDHQLR | LLEAQKRNQE | VVLRRKTEEV | TALRRQVRPM |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SDKVAGKVTR | KLSSSDAPAQ | DTGSSAAAVE | TDASRTGAQQ | KMRIPVARVQ | ALPTPATNGN |
| 910 | 920 | 930 | 940 | 950 | 960 |
| RKKYQRKGLT | GRVFISKTAR | MKWQLLERRV | TDIIMQKMTI | SNMEADMNRL | LKQREELTKR |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| REKLSKRREK | IVKENGEGDK | NVANINEEME | SLTANIDYIN | DSISDCQANI | MQMEEAKEEG |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| ETLDVTAVIN | ACTLTEARYL | LDHFLSMGIN | KGLQAAQKEA | QIKVLEGRLK | QTEITSATQN |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| QLLFHMLKEK | AELNPELDAL | LGHALQDLDS | VPLENVEDST | DEDAPLNSPG | SEGSTLSSDL |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| MKLCGEVKPK | NKARRRTTTQ | MELLYADSSE | LASDTSTGDA | SLPGPLTPVA | EGQEIGMNTE |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| TSGTSAREKE | LSPPPGLPSK | IGSISRQSSL | SEKKIPEPSP | VTRRKAYEKA | EKSKAKEQKH |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| SDSGTSEASL | SPPSSPPSRP | RNELNVFNRL | TVSQGNTSVQ | QDKSDESDSS | LSEVHRSSRR |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| GIINPFPASK | GIRAFPLQCI | HIAEGHTKAV | LCVDSTDDLL | FTGSKDRTCK | VWNLVTGQEI |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| MSLGGHPNNV | VSVKYCNYTS | LVFTVSTSYI | KVWDIRDSAK | CIRTLTSSGQ | VTLGDACSAS |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| TSRTVAIPSG | ENQINQIALN | PTGTFLYAAS | GNAVRMWDLK | RFQSTGKLTG | HLGPVMCLTV |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| DQISSGQDLI | ITGSKDHYIK | MFDVTEGALG | TVSPTHNFEP | PHYDGIEALT | IQGDNLFSGS |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| RDNGIKKWDL | TQKDLLQQVP | NAHKDWVCAL | GVVPDHPVLL | SGCRGGILKV | WNMDTFMPVG |
| 1630 | 1640 | 1650 | 1660 | 1670 | |
| EMKGHDSPIN | AICVNSTHIF | TAADDRTVRI | WKARNLQDGQ | ISDTGDLGED | IASN |