Q7YRV4
Gene name |
TRIM21 |
Protein name |
E3 ubiquitin-protein ligase TRIM21 |
Names |
52 kDa Ro protein, 52 kDa ribonucleoprotein autoantigen Ro/SS-A, RING-type E3 ubiquitin transferase TRIM21, Ro(SS-A), Sjoegren syndrome type A antigen, SS-A, Tripartite motif-containing protein 21 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:359715 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
|
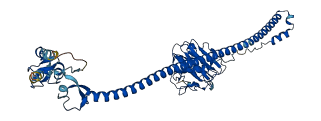
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q7YRV4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q7YRV4-F1 | Predicted | AlphaFoldDB |
85 variants for Q7YRV4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs479813467 | 8 | T>P | No | EVA | |
| rs457566432 | 91 | L>M | No | EVA | |
| rs469544882 | 95 | H>Q | No | EVA | |
| rs433485035 | 100 | H>P | No | EVA | |
| rs451976534 | 110 | L>P | No | EVA | |
| rs472985840 | 113 | V>L | No | EVA | |
| rs455242855 | 115 | S>P | No | EVA | |
| rs473958747 | 119 | K>N | No | EVA | |
| rs444057682 | 120 | H>P | No | EVA | |
| rs441292125 | 121 | R>H | No | EVA | |
| rs471362794 | 122 | D>E | No | EVA | |
| rs462606057 | 122 | D>V | No | EVA | |
| rs460313784 | 123 | H>R | No | EVA | |
| rs438668238 | 123 | H>Y | No | EVA | |
| rs462107712 | 128 | I>L | No | EVA | |
| rs450994202 | 130 | E>G | No | EVA | |
| rs480768746 | 130 | E>K | No | EVA | |
| rs433316228 | 132 | A>S | No | EVA | |
| rs445449040 | 133 | Q>* | No | EVA | |
| rs467338103 | 133 | Q>P | No | EVA | |
| rs455340153 | 134 | E>D | No | EVA | |
| rs434255577 | 134 | E>G | No | EVA | |
| rs473773092 | 136 | Q>P | No | EVA | |
| rs478336936 | 167 | W>* | No | EVA | |
| rs438893744 | 168 | K>M | No | EVA | |
| rs460755389 | 168 | K>N | No | EVA | |
| rs479779565 | 182 | F>V | No | EVA | |
| rs436434188 | 210 | T>A | No | EVA | |
| rs454774869 | 214 | T>P | No | EVA | |
| rs470140553 | 214 | T>R | No | EVA | |
| rs452607844 | 218 | L>V | No | EVA | |
| rs471180084 | 219 | A>G | No | EVA | |
| rs876359373 | 224 | A>V | No | EVA | |
| rs876422182 | 225 | L>P | No | EVA | |
| rs453624882 | 225 | L>V | No | EVA | |
| rs480097732 | 226 | Q>H | No | EVA | |
| rs475301551 | 226 | Q>L | No | EVA | |
| rs441665048 | 228 | L>M | No | EVA | |
| rs876270569 | 240 | A>G | No | EVA | |
| rs463334803 | 241 | L>R | No | EVA | |
| rs481931109 | 242 | E>D | No | EVA | |
| rs875990097 | 242 | E>G | No | EVA | |
| rs876224705 | 243 | L>V | No | EVA | |
| rs439296603 | 244 | L>P | No | EVA | |
| rs876286743 | 244 | L>V | No | EVA | |
| rs876021808 | 245 | Q>E | No | EVA | |
| rs876031495 | 245 | Q>H | No | EVA | |
| rs876080029 | 245 | Q>P | No | EVA | |
| rs132673159 | 256 | S>F | No | EVA | |
| rs456489148 | 287 | V>E | No | EVA | |
| rs432483258 | 288 | H>P | No | EVA | |
| rs454247783 | 290 | T>A | No | EVA | |
| rs443005209 | 294 | Q>* | No | EVA | |
| rs379823616 | 306 | R>Q | No | EVA | |
| rs476724384 | 312 | A>S | No | EVA | |
| rs443891041 | 313 | N>K | No | EVA | |
| rs476998777 | 316 | Q>K | No | EVA | |
| rs440787314 | 318 | V>G | No | EVA | |
| rs448303333 | 323 | E>D | No | EVA | |
| rs481092145 | 323 | E>G | No | EVA | |
| rs482117037 | 325 | F>V | No | EVA | |
| rs445879505 | 326 | D>G | No | EVA | |
| rs464413683 | 328 | Y>F | No | EVA | |
| rs464413683 | 328 | Y>S | No | EVA | |
| rs454020755 | 331 | V>G | No | EVA | |
| rs454997771 | 338 | D>E | No | EVA | |
| rs715374961 | 339 | S>C | No | EVA | |
| rs443902779 | 348 | V>E | No | EVA | |
| rs452815616 | 349 | T>M | No | EVA | |
| rs440819537 | 351 | K>R | No | EVA | |
| rs444742169 | 354 | W>* | No | EVA | |
| rs481132856 | 359 | C>Y | No | EVA | |
| rs463556270 | 370 | L>M | No | EVA | |
| rs135296314 | 385 | K>R | No | EVA | |
| rs445789279 | 392 | T>P | No | EVA | |
| rs464492631 | 405 | N>H | No | EVA | |
| rs479742605 | 405 | N>T | No | EVA | |
| rs447612822 | 408 | G>E | No | EVA | |
| rs436457657 | 414 | E>K | No | EVA | |
| rs448453870 | 418 | V>G | No | EVA | |
| rs437440579 | 423 | I>S | No | EVA | |
| rs452602128 | 428 | S>P | No | EVA | |
| rs435047248 | 430 | I>L | No | EVA | |
| rs452927526 | 455 | T>A | No | EVA | |
| rs474555546 | 457 | S>P | No | EVA |
No associated diseases with Q7YRV4
1 regional properties for Q7YRV4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | MATH/TRAF domain | 6 - 137 | IPR002083 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| autophagosome | A double-membrane-bounded compartment that engulfs endogenous cellular material as well as invading microorganisms to target them to the lytic vacuole/lysosome for degradation as part of macroautophagy. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| P-body | A focus in the cytoplasm where mRNAs may become inactivated by decapping or some other mechanism. Protein and RNA localized to these foci are involved in mRNA degradation, nonsense-mediated mRNA decay (NMD), translational repression, and RNA-mediated gene silencing. |
| ribonucleoprotein complex | A macromolecular complex that contains both RNA and protein molecules. |
| SCF ubiquitin ligase complex | A ubiquitin ligase complex in which a cullin from the Cul1 subfamily and a RING domain protein form the catalytic core; substrate specificity is conferred by a Skp1 adaptor and an F-box protein. SCF complexes are involved in targeting proteins for degradation by the proteasome. The best characterized complexes are those from yeast and mammals (with core subunits named Cdc53/Cul1, Rbx1/Hrt1/Roc1). |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| transcription coactivator activity | A transcription coregulator activity that activates or increases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Coactivators often act by altering chromatin structure and modifications. For example, one class of transcription coactivators modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. A fourth class of coactivator activity is the bridging of a DNA-binding transcription factor to the general (basal) transcription machinery. The Mediator complex, which bridges sequence-specific DNA binding transcription factors and RNA polymerase, is also a transcription coactivator. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
| zinc ion binding | Binding to a zinc ion (Zn). |
23 GO annotations of biological process
| Name | Definition |
|---|---|
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| negative regulation of innate immune response | Any process that stops, prevents, or reduces the frequency, rate or extent of the innate immune response. |
| negative regulation of NF-kappaB transcription factor activity | Any process that stops, prevents, or reduces the frequency, rate or extent of the activity of the transcription factor NF-kappaB. |
| negative regulation of protein deubiquitination | Any process that decreases the frequency, rate or extent of protein deubiquitination. Protein deubiquitination is the removal of one or more ubiquitin groups from a protein. |
| negative regulation of viral transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of viral transcription. |
| positive regulation of autophagy | Any process that activates, maintains or increases the rate of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| positive regulation of cell cycle | Any process that activates or increases the rate or extent of progression through the cell cycle. |
| positive regulation of I-kappaB kinase/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| positive regulation of NF-kappaB transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of the transcription factor NF-kappaB. |
| positive regulation of protein binding | Any process that activates or increases the frequency, rate or extent of protein binding. |
| positive regulation of viral entry into host cell | Any process that activates or increases the frequency, rate or extent of the introduction of viral entry into the host cell. |
| protein autoubiquitination | The ubiquitination by a protein of one or more of its own amino acid residues, or residues on an identical protein. Ubiquitination occurs on the lysine residue by formation of an isopeptide crosslink. |
| protein destabilization | Any process that decreases the stability of a protein, making it more vulnerable to degradative processes or aggregation. |
| protein K63-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 63 of the ubiquitin monomers, is added to a protein. K63-linked ubiquitination does not target the substrate protein for degradation, but is involved in several pathways, notably as a signal to promote error-free DNA postreplication repair. |
| protein monoubiquitination | Addition of a single ubiquitin group to a protein. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of protein localization | Any process that modulates the frequency, rate or extent of any process in which a protein is transported to, or maintained in, a specific location. |
| regulation of viral entry into host cell | Any process that modulates the frequency, rate or extent of the viral entry into the host cell. |
| response to interferon-gamma | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interferon-gamma stimulus. Interferon-gamma is also known as type II interferon. |
| suppression of viral release by host | A process in which a host organism stops, prevents or reduces the frequency, rate or extent of the release of a virus with which it is infected, from its cells. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| E1BJS7 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Bos taurus (Bovine) | PR |
| Q5E9G4 | TRIM10 | Tripartite motif-containing protein 10 | Bos taurus (Bovine) | PR |
| Q2T9Z0 | TRIM17 | E3 ubiquitin-protein ligase TRIM17 | Bos taurus (Bovine) | PR |
| Q1PRL4 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Gallus gallus (Chicken) | PR |
| Q7YR32 | TRIM10 | Tripartite motif-containing protein 10 | Pan troglodytes (Chimpanzee) | PR |
| O15553 | MEFV | Pyrin | Homo sapiens (Human) | SS |
| Q9BTV5 | FSD1 | Fibronectin type III and SPRY domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q9H2S5 | RNF39 | RING finger protein 39 | Homo sapiens (Human) | PR |
| Q9UJV3 | MID2 | Probable E3 ubiquitin-protein ligase MID2 | Homo sapiens (Human) | PR |
| P29590 | PML | Protein PML | Homo sapiens (Human) | PR |
| Q9C029 | TRIM7 | E3 ubiquitin-protein ligase TRIM7 | Homo sapiens (Human) | PR |
| Q9NQ86 | TRIM36 | E3 ubiquitin-protein ligase TRIM36 | Homo sapiens (Human) | PR |
| Q86UV6 | TRIM74 | Tripartite motif-containing protein 74 | Homo sapiens (Human) | PR |
| Q9UPQ4 | TRIM35 | E3 ubiquitin-protein ligase TRIM35 | Homo sapiens (Human) | PR |
| Q6ZMU5 | TRIM72 | Tripartite motif-containing protein 72 | Homo sapiens (Human) | PR |
| Q86UV7 | TRIM73 | Tripartite motif-containing protein 73 | Homo sapiens (Human) | PR |
| Q8N9V2 | TRIML1 | Probable E3 ubiquitin-protein ligase TRIML1 | Homo sapiens (Human) | PR |
| Q86XT4 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Homo sapiens (Human) | PR |
| Q5EBN2 | TRIM61 | Putative tripartite motif-containing protein 61 | Homo sapiens (Human) | PR |
| Q9BZY9 | TRIM31 | E3 ubiquitin-protein ligase TRIM31 | Homo sapiens (Human) | PR |
| Q2Q1W2 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Homo sapiens (Human) | PR |
| Q9BXM9 | FSD1L | FSD1-like protein | Homo sapiens (Human) | PR |
| P19474 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Homo sapiens (Human) | PR |
| Q9WUH5 | Trim10 | Tripartite motif-containing protein 10 | Mus musculus (Mouse) | PR |
| Q8BZT2 | Sh3rf2 | E3 ubiquitin-protein ligase SH3RF2 | Mus musculus (Mouse) | PR |
| Q7TPM3 | Trim17 | E3 ubiquitin-protein ligase TRIM17 | Mus musculus (Mouse) | PR |
| Q1XH17 | Trim72 | Tripartite motif-containing protein 72 | Mus musculus (Mouse) | PR |
| Q60953 | Pml | Protein PML | Mus musculus (Mouse) | PR |
| Q9JJ26 | Mefv | Pyrin | Mus musculus (Mouse) | SS |
| Q99PQ1 | Trim12a | Tripartite motif-containing protein 12A | Mus musculus (Mouse) | PR |
| Q61510 | Trim25 | E3 ubiquitin/ISG15 ligase TRIM25 | Mus musculus (Mouse) | PR |
| Q810I2 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Mus musculus (Mouse) | PR |
| Q1PSW8 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Mus musculus (Mouse) | PR |
| Q3TL54 | Trim43a | Tripartite motif-containing protein 43A | Mus musculus (Mouse) | PR |
| P86449 | Trim43c | Tripartite motif-containing protein 43C | Mus musculus (Mouse) | PR |
| O77666 | TRIM26 | Tripartite motif-containing protein 26 | Sus scrofa (Pig) | PR |
| O19085 | TRIM10 | Tripartite motif-containing protein 10 | Sus scrofa (Pig) | PR |
| Q865W2 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Sus scrofa (Pig) | PR |
| Q920M2 | Rnf39 | RING finger protein 39 | Rattus norvegicus (Rat) | PR |
| Q9JJ25 | Mefv | Pyrin | Rattus norvegicus (Rat) | SS |
| A0JPQ4 | Trim72 | Tripartite motif-containing protein 72 | Rattus norvegicus (Rat) | PR |
| Q810I1 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Rattus norvegicus (Rat) | PR |
| D3ZVM4 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Rattus norvegicus (Rat) | PR |
| F6QEU4 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q640S6 | trim72 | Tripartite motif-containing protein 72 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| E7FAM5 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASAVPLTMM | WEEVTCSICL | DPMVEPMSIE | CGHSFCQECI | SEVGKEGGSV | CPVCRRHFLL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QNLRPNRQVA | NMVDNLRKIS | QGAKESPHGE | LCVVHREKIH | LFCEEDGKAL | CWVCSQSQKH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RDHPMVPIEE | AAQEYQEKLQ | VALNKLRHKQ | ELAEKLELDI | AMKKASWKGK | VEAHKLRIHE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EFVRQKNFLA | EEEQKQLQKL | EEEERQQLRT | LGDTEARLAQ | QIQALQELIA | ELERRRRGSA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LELLQEVKSV | LERSESWNLK | ELDVASTDLR | NVFYVPGIKK | MLRTCGVHIT | LDPQTANPWL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ILSENRRQVR | LANTRQEVPE | NEERFDSYPM | VLGAQRFDSG | KVYWEVDVTG | KEAWDLGVCR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DNVRRKGQFL | LNSDTGFWII | WLWNKQKYEA | GTCPQTPLHL | QVPPNRIGIF | LDYEASTVSF |
| 430 | 440 | 450 | 460 | ||
| YNITDHASLI | YTFSECAFAG | PLRPFFNPGF | NDRGTNSAPL | ILCPLKTGW |