Q7X8C5
Gene name |
WAKL2 (At1g16130, T24D18.21) |
Protein name |
Wall-associated receptor kinase-like 2 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G16130 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
558-582 (Activation loop from InterPro)
Target domain |
416-689 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
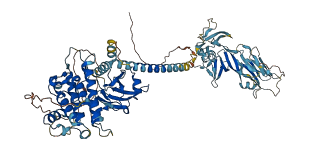
Autoinhibited structure

Activated structure

1 structures for Q7X8C5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q7X8C5-F1 | Predicted | AlphaFoldDB |
43 variants for Q7X8C5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH04584431 | 12 | P>L | No | 1000Genomes | |
| tmp_1_5525674_C_G | 14 | A>G | No | 1000Genomes | |
| tmp_1_5525716_C_T | 28 | A>V | No | 1000Genomes | |
| ENSVATH01057537 | 91 | H>P | No | 1000Genomes | |
| ENSVATH11312313 | 100 | G>S | No | 1000Genomes | |
| tmp_1_5525962_C_G | 110 | T>R | No | 1000Genomes | |
| ENSVATH01057538 | 111 | P>S | No | 1000Genomes | |
| tmp_1_5525992_G_T | 120 | G>V | No | 1000Genomes | |
| tmp_1_5525994_A_T | 121 | S>C | No | 1000Genomes | |
| ENSVATH04584434 | 132 | V>L | No | 1000Genomes | |
| ENSVATH01057539 | 161 | S>N | No | 1000Genomes | |
| ENSVATH01057541 | 235 | A>V | No | 1000Genomes | |
| ENSVATH11312431 | 277 | F>L | No | 1000Genomes | |
| ENSVATH01057542 | 308 | C>Y | No | 1000Genomes | |
| ENSVATH01057543 | 315 | N>H | No | 1000Genomes | |
| tmp_1_5526953_A_G | 385 | M>V | No | 1000Genomes | |
| tmp_1_5526991_T_A | 397 | N>K | No | 1000Genomes | |
| tmp_1_5527052_A_C | 418 | K>Q | No | 1000Genomes | |
| tmp_1_5527070_C_A | 424 | Q>K | No | 1000Genomes | |
| ENSVATH13904857 | 427 | Q>* | No | 1000Genomes | |
| ENSVATH04584440 | 445 | R>M | No | 1000Genomes | |
| tmp_1_5527256_G_A | 486 | V>I | No | 1000Genomes | |
| tmp_1_5527277_C_T | 493 | P>S | No | 1000Genomes | |
| ENSVATH11312432 | 511 | T>K | No | 1000Genomes | |
| tmp_1_5527373_G_A | 525 | A>T | No | 1000Genomes | |
| ENSVATH13904858 | 559 | D>G | No | 1000Genomes | |
| ENSVATH11312434 | 559 | D>N | No | 1000Genomes | |
| tmp_1_5527503_T_A | 568 | I>K | No | 1000Genomes | |
| ENSVATH11312545 | 569 | D>E | No | 1000Genomes | |
| ENSVATH11312546 | 575 | T>I | No | 1000Genomes | |
| tmp_1_5527602_A_T | 601 | Y>F | No | 1000Genomes | |
| tmp_1_5527635_C_T | 612 | T>I | No | 1000Genomes | |
| tmp_1_5527655_C_T | 619 | R>C | No | 1000Genomes | |
| tmp_1_5527674_A_G | 625 | N>S | No | 1000Genomes | |
| tmp_1_5527694_T_G | 632 | F>V | No | 1000Genomes | |
| ENSVATH04584441 | 648 | R>Q | No | 1000Genomes | |
| ENSVATH11312554 | 700 | I>T | No | 1000Genomes | |
| ENSVATH11312625 | 703 | D>Y | No | 1000Genomes | |
| ENSVATH01057546 | 710 | A>V | No | 1000Genomes | |
| tmp_1_5527937_C_G | 713 | L>V | No | 1000Genomes | |
| ENSVATH11312627 | 722 | G>C | No | 1000Genomes | |
| ENSVATH00026238 | 723 | A>V | No | 1000Genomes | |
| ENSVATH11312628 | 729 | M>T | No | 1000Genomes |
No associated diseases with Q7X8C5
5 regional properties for Q7X8C5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 416 - 689 | IPR000719 |
| active_site | Serine/threonine-protein kinase, active site | 537 - 549 | IPR008271 |
| domain | Wall-associated receptor kinase | 168 - 274 | IPR013695 |
| conserved_site | EGF-like calcium-binding, conserved site | 304 - 331 | IPR018097 |
| domain | Wall-associated receptor kinase, galacturonan-binding domain | 35 - 86 | IPR025287 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plant-type cell wall | A more or less rigid stucture lying outside the cell membrane of a cell and composed of cellulose and pectin and other organic and inorganic substances. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| polysaccharide binding | Binding to a polysaccharide, a polymer of many (typically more than 10) monosaccharide residues linked glycosidically. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| P06213 | INSR | Insulin receptor | Homo sapiens (Human) | EV |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | Homo sapiens (Human) | EV |
| P14616 | INSRR | Insulin receptor-related protein | Homo sapiens (Human) | SS |
| P00533 | EGFR | Epidermal growth factor receptor | Homo sapiens (Human) | EV |
| Q15303 | ERBB4 | Receptor tyrosine-protein kinase erbB-4 | Homo sapiens (Human) | SS |
| P04626 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | Homo sapiens (Human) | SS |
| P21860 | ERBB3 | Receptor tyrosine-protein kinase erbB-3 | Homo sapiens (Human) | SS |
| P05532 | Kit | Mast/stem cell growth factor receptor Kit | Mus musculus (Mouse) | PR |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| P20786 | Pdgfra | Platelet-derived growth factor receptor alpha | Rattus norvegicus (Rat) | SS |
| Q05030 | Pdgfrb | Platelet-derived growth factor receptor beta | Rattus norvegicus (Rat) | SS |
| O08775 | Kdr | Vascular endothelial growth factor receptor 2 | Rattus norvegicus (Rat) | SS |
| Q498D6 | Fgfr4 | Fibroblast growth factor receptor 4 | Rattus norvegicus (Rat) | PR |
| Q04589 | Fgfr1 | Fibroblast growth factor receptor 1 | Rattus norvegicus (Rat) | SS |
| Q00495 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Rattus norvegicus (Rat) | PR |
| Q8VYA3 | WAKL10 | Wall-associated receptor kinase-like 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C9L5 | WAKL9 | Wall-associated receptor kinase-like 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M092 | WAKL17 | Wall-associated receptor kinase-like 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9S9M2 | WAKL4 | Wall-associated receptor kinase-like 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9YI66 | tyro3 | Tyrosine-protein kinase receptor TYRO3 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKTETHNRQC | IPLAISVLSL | FINGVSSARQ | PPDRCNRVCG | EISIPFPFGI | GGKDCYLNPW |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YEVVCNSTNS | VPFLSRINRE | LVNISLNGVV | HIKAPVTSSG | CSTGTSQPLT | PPPLNVAGQG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SPYFLTDKNL | LVAVGCKFKA | VMAGITSQIT | SCESSCNERN | SSSQEGRNKI | CNGYKCCQTR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IPEGQPQVIS | VDIEIPQGNN | TTGEGGCRVA | FLTSDKYSSL | NVTEPEKFHG | HGYAAVELGW |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FFDTSDSRDT | QPISCKNASD | TTPYTSDTRC | SCSYGYFSGF | SYRDCYCNSP | GYKGNPFLPG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GCVDVDECKL | DIGRNQCKDQ | SCVNLPGWFD | CQPKKPEQLK | RVIQGVLIGS | ALLLFAFGIF |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GLYKFVQKRR | KLIRMRKFFR | RNGGMLLKQQ | LARKEGNVEM | SRIFSSHELE | KATDNFNKNR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VLGQGGQGTV | YKGMLVDGRI | VAVKRSKAVD | EDRVEEFINE | VVVLAQINHR | NIVKLLGCCL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ETEVPVLVYE | FVPNGDLCKR | LHDESDDYTM | TWEVRLHIAI | EIAGALSYLH | SAASFPIYHR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DIKTTNILLD | ERNRAKVSDF | GTSRSVTIDQ | THLTTQVAGT | FGYVDPEYFQ | SSKFTEKSDV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| YSFGVVLVEL | LTGEKPSSRV | RSEENRGLAA | HFVEAVKENR | VLDIVDDRIK | DECNMDQVMS |
| 670 | 680 | 690 | 700 | 710 | 720 |
| VANLARRCLN | RKGKKRPNMR | EVSIELEMIR | SSHYDSGIHI | EDDDEEDDQA | MELNFNDTWE |
| 730 | 740 | ||||
| VGATAPASMF | NNASPTSDAE | PLVPLRTW |