Q7TT36
Gene name |
Adgra3 (Gpr125) |
Protein name |
Adhesion G protein-coupled receptor A3 |
Names |
G-protein coupled receptor 125 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:70693 |
EC number |
|
Protein Class |
|
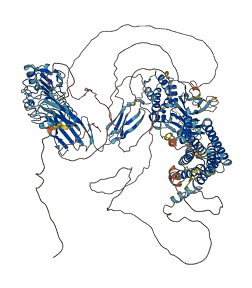
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q7TT36
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q7TT36-F1 | Predicted | AlphaFoldDB |
71 variants for Q7TT36
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3395180331 | 4 | P>L | No | EVA | |
| rs3395557580 | 28 | L>Q | No | EVA | |
| rs3388732446 | 80 | N>S | No | EVA | |
| rs3388732484 | 82 | I>N | No | EVA | |
| rs3388743738 | 88 | G>A | No | EVA | |
| rs3388761159 | 101 | L>F | No | EVA | |
| rs3388761132 | 102 | R>G | No | EVA | |
| rs3388758307 | 110 | A>G | No | EVA | |
| rs3388743786 | 137 | V>E | No | EVA | |
| rs3395502442 | 183 | N>D | No | EVA | |
| rs263503117 | 189 | R>G | No | EVA | |
| rs3388732453 | 271 | R>S | No | EVA | |
| rs3388758273 | 329 | V>L | No | EVA | |
| rs3388760261 | 356 | L>Q | No | EVA | |
| rs3388760244 | 393 | G>E | No | EVA | |
| rs3388760295 | 401 | S>C | No | EVA | |
| rs3388760598 | 411 | R>K | No | EVA | |
| rs3388764527 | 415 | M>I | No | EVA | |
| rs253622930 | 423 | L>V | No | EVA | |
| rs3388761833 | 477 | D>G | No | EVA | |
| rs3388764514 | 488 | R>Q | No | EVA | |
| rs3388758234 | 501 | S>N | No | EVA | |
| rs3388753368 | 506 | C>S | No | EVA | |
| rs3388756928 | 508 | Q>* | No | EVA | |
| rs3388761129 | 513 | H>L | No | EVA | |
| rs3388743799 | 571 | N>K | No | EVA | |
| rs244191463 | 611 | H>P | No | EVA | |
| rs223511038 | 614 | E>D | No | EVA | |
| rs235460781 | 673 | I>V | No | EVA | |
| rs255284624 | 724 | G>S | No | EVA | |
| rs3388760245 | 806 | T>N | No | EVA | |
| rs3395314501 | 829 | V>I | No | EVA | |
| rs3395385719 | 830 | L>S | No | EVA | |
| rs3388761111 | 849 | R>I | No | EVA | |
| rs3388753347 | 871 | G>D | No | EVA | |
| rs3388761811 | 906 | L>W | No | EVA | |
| rs3388750222 | 914 | S>I | No | EVA | |
| rs3388761875 | 914 | S>R | No | EVA | |
| rs3388753299 | 923 | Y>* | No | EVA | |
| rs3388761812 | 953 | A>D | No | EVA | |
| rs3388753309 | 974 | T>M | No | EVA | |
| rs235087732 | 976 | E>D | No | EVA | |
| rs3388750216 | 978 | E>D | No | EVA | |
| rs3388761162 | 993 | L>P | No | EVA | |
| rs257395133 | 997 | I>T | No | EVA | |
| rs3388764570 | 1022 | A>D | No | EVA | |
| rs3388745826 | 1037 | I>F | No | EVA | |
| rs3388766908 | 1044 | L>F | No | EVA | |
| rs248932780 | 1049 | M>L | No | EVA | |
| rs3388757847 | 1059 | V>M | No | EVA | |
| rs3388760546 | 1064 | Q>K | No | EVA | |
| rs32908609 | 1070 | A>T | No | EVA | |
| rs32908611 | 1097 | N>T | No | EVA | |
| rs3412243573 | 1101 | G>D | No | EVA | |
| rs3413134851 | 1102 | C>R | No | EVA | |
| rs3388745807 | 1113 | Q>* | No | EVA | |
| rs3388757846 | 1118 | A>G | No | EVA | |
| rs3388750194 | 1124 | T>M | No | EVA | |
| rs3388766903 | 1161 | H>N | No | EVA | |
| rs3388760291 | 1163 | N>Y | No | EVA | |
| rs3388760622 | 1175 | V>F | No | EVA | |
| rs3388761873 | 1198 | S>G | No | EVA | |
| rs3388761200 | 1200 | P>L | No | EVA | |
| rs3388760325 | 1207 | S>* | No | EVA | |
| rs3388757913 | 1245 | P>H | No | EVA | |
| rs3388764534 | 1252 | K>R | No | EVA | |
| rs3388761165 | 1273 | N>I | No | EVA | |
| rs3388766891 | 1283 | S>G | No | EVA | |
| rs3395179155 | 1286 | Q>L | No | EVA | |
| rs3395505853 | 1294 | D>S | No | EVA | |
| rs3388766895 | 1304 | W>R | No | EVA |
No associated diseases with Q7TT36
12 regional properties for Q7TT36
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | GPS motif | 686 - 738 | IPR000203 |
| domain | Cysteine-rich flanking region, C-terminal | 176 - 225 | IPR000483 |
| repeat | Leucine-rich repeat | 74 - 130 | IPR001611 |
| domain | GPCR, family 2, extracellular hormone receptor domain | 312 - 407 | IPR001879 |
| repeat | Leucine-rich repeat, typical subtype | 68 - 92 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 93 - 116 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 117 - 140 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 141 - 164 | IPR003591-4 |
| domain | Immunoglobulin subtype | 238 - 331 | IPR003599 |
| domain | Immunoglobulin-like domain | 231 - 329 | IPR007110 |
| domain | Immunoglobulin I-set | 236 - 329 | IPR013098 |
| domain | GPCR, family 2-like, transmembrane domain | 745 - 1039 | IPR017981 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| external side of plasma membrane | The leaflet of the plasma membrane that faces away from the cytoplasm and any proteins embedded or anchored in it or attached to its surface. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein-coupled receptor activity | Combining with an extracellular signal and transmitting the signal across the membrane by activating an associated G-protein; promotes the exchange of GDP for GTP on the alpha subunit of a heterotrimeric G-protein complex. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
39 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3MHH9 | ECM2 | Extracellular matrix protein 2 | Bos taurus (Bovine) | PR |
| P58874 | OPTC | Opticin | Bos taurus (Bovine) | PR |
| Q24K06 | LRRC10 | Leucine-rich repeat-containing protein 10 | Bos taurus (Bovine) | PR |
| A6H789 | LRRC3B | Leucine-rich repeat-containing protein 3B | Bos taurus (Bovine) | PR |
| Q9V780 | Lap1 | Protein lap1 | Drosophila melanogaster (Fruit fly) | PR |
| Q96NW7 | LRRC7 | Leucine-rich repeat-containing protein 7 | Homo sapiens (Human) | PR |
| Q8IWT6 | LRRC8A | Volume-regulated anion channel subunit LRRC8A | Homo sapiens (Human) | PR |
| Q9HCJ2 | LRRC4C | Leucine-rich repeat-containing protein 4C | Homo sapiens (Human) | PR |
| Q9UFC0 | LRWD1 | Leucine-rich repeat and WD repeat-containing protein 1 | Homo sapiens (Human) | PR |
| Q96L50 | LRR1 | Leucine-rich repeat protein 1 | Homo sapiens (Human) | PR |
| Q86UN2 | RTN4RL1 | Reticulon-4 receptor-like 1 | Homo sapiens (Human) | PR |
| Q7L1W4 | LRRC8D | Volume-regulated anion channel subunit LRRC8D | Homo sapiens (Human) | PR |
| Q96FE5 | LINGO1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Homo sapiens (Human) | PR |
| Q8TDW0 | LRRC8C | Volume-regulated anion channel subunit LRRC8C | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| Q96PB8 | LRRC3B | Leucine-rich repeat-containing protein 3B | Homo sapiens (Human) | PR |
| Q8IWK6 | ADGRA3 | Adhesion G protein-coupled receptor A3 | Homo sapiens (Human) | PR |
| A6H694 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Mus musculus (Mouse) | PR |
| Q9D9Q0 | Lrrc69 | Leucine-rich repeat-containing protein 69 | Mus musculus (Mouse) | PR |
| Q8BGI7 | Lrrc39 | Leucine-rich repeat-containing protein 39 | Mus musculus (Mouse) | PR |
| P59383 | Lrrn4 | Leucine-rich repeat neuronal protein 4 | Mus musculus (Mouse) | PR |
| Q9D1T0 | Lingo1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Mus musculus (Mouse) | PR |
| Q8VCH9 | Lrrc3b | Leucine-rich repeat-containing protein 3B | Mus musculus (Mouse) | PR |
| Q8R502 | Lrrc8c | Volume-regulated anion channel subunit LRRC8C | Mus musculus (Mouse) | PR |
| Q80WG5 | Lrrc8a | Volume-regulated anion channel subunit LRRC8A | Mus musculus (Mouse) | PR |
| Q8K0S5 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Mus musculus (Mouse) | PR |
| Q80TE7 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Mus musculus (Mouse) | PR |
| Q5DU41 | Lrrc8b | Volume-regulated anion channel subunit LRRC8B | Mus musculus (Mouse) | PR |
| Q5RKR3 | Islr2 | Immunoglobulin superfamily containing leucine-rich repeat protein 2 | Mus musculus (Mouse) | PR |
| Q8C031 | Lrrc4c | Leucine-rich repeat-containing protein 4C | Mus musculus (Mouse) | PR |
| P70587 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Rattus norvegicus (Rat) | PR |
| Q4V8G0 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Rattus norvegicus (Rat) | PR |
| Q80WD0 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Rattus norvegicus (Rat) | PR |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9SHI4 | RLP3 | Receptor-like protein 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5G5E0 | PIRL5 | Plant intracellular Ras-group-related LRR protein 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| B0JZ65 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q68F79 | lrrc8e | Volume-regulated anion channel subunit LRRC8E | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| B0R160 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEPPPPLLLL | PLALLALLWG | GERGAAALPA | GCKHDGRARG | TGRAAAAAEG | KVVCSSLELA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QVLPPDTLPN | RTVTLILSNN | KISELKNGSF | SGLSLLERLD | LRNNLISRIA | PGAFWGLSSL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KRLDLTNNRI | GCLNADVFRG | LTNLVRLNLS | GNLFTSLSQG | TFDYLGSLRS | LEFQTEYLLC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DCNILWMHRW | VKERNITVRD | TRCVYPKSLQ | AQPVTGVKQE | LLTCDPPLEL | PSFYMTPSHR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QVVFEGDSLP | FQCMASYIDQ | DMQVLWYQDG | RIVETDESQG | IFVEKSMIHN | CSLIASALTI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SNIQAGSTGN | WGCHVQTKRG | NNTRTVDIVV | LESSAQYCPP | ERVVNNKGDF | RWPRTLAGIT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AYLQCTRNTH | SSGIYPGSAQ | DERKAWRRCD | RGGFWADDDY | SRCQYANDVT | RVLYMFNQMP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LNLTNAVATA | RQLLAYTVEA | ANFSDKMDVI | FVAEMIEKFG | RFTREEKSKE | LGDVMVDVAS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| NIMLADERVL | WLAQREAKAC | SRIVQCLQRI | ATHRLASGAH | VYSTYSPNIA | LEAYVIKAAG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| FTGMTCSVFQ | KVAASDRAGL | SDYGRRDPDG | NLDKQLSFKC | NVSSTFSSLA | LKNTIMEASI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| QLPSSLLSPK | HKREARAADD | ALYKLQLIAF | RNGKLFPATG | NSTKLADDGK | RRTVVTPVIL |
| 670 | 680 | 690 | 700 | 710 | 720 |
| TKIDGATVDT | HHIPVNVTLR | RIAHGADAVA | AQWDFDLLNG | QGGWKSDGCC | ILYSDENITT |
| 730 | 740 | 750 | 760 | 770 | 780 |
| IQCGSLGNYA | VLMDLTGTEL | YTPAASLLHP | VVYTTAITLL | LCLLAVIISY | MYHHSLIRIS |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LKSWHMLVNL | CFHILLTCVV | FVGGITQTRN | ASVCQAVGII | LHYSTLATVL | WVGVTARNIY |
| 850 | 860 | 870 | 880 | 890 | 900 |
| KQVTKKAKRC | QDPDEPPAPP | RPMLRFYLIG | GGIPIIVCGI | TAAANIKNYG | SRPSAPYCWM |
| 910 | 920 | 930 | 940 | 950 | 960 |
| AWEPSLGAFY | GPASFITFVN | CMYFLSIFIQ | LKRHPERKYE | LKEPTEEQQR | LAANENGEIN |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| HQDSMSLSLI | STSTLENEHS | FQSQLLGASL | TLLLYVILWM | FGAMAVSLYY | PLDLVFSFFF |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| GATCLSFSAF | MMVHHCINRE | DVRLAWIMMC | CPGRSSYSVQ | VNVQPPNSSA | TNGEAPKCTN |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| SSAESSCTNK | SASSFKNSSQ | GCKLTNLQAA | AAQYHSNALP | VNATPQLDNS | LTEHSMDNDI |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| KMHVAPLDVQ | FRTNVHPSRH | HKNRSKGHRA | SRLTVLREYA | YDVPTSVEGS | VQNGLPKSRP |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| GSNEGHSRSR | RAYLAYRERQ | YNPPQQDSSD | ACSTLPKSSR | NVEKPVSTSS | KKDAPRKPAA |
| 1270 | 1280 | 1290 | 1300 | ||
| ADLESQQKSY | GLNLAVQNGP | VKSNGQEGPL | LATDVTGNVR | TGLWKHETTV |