Q7SXW3
Gene name |
lrrc40 (zgc:63729) |
Protein name |
Leucine-rich repeat-containing protein 40 |
Names |
|
Species |
Danio rerio (Zebrafish) (Brachydanio rerio) |
KEGG Pathway |
dre:334130 |
EC number |
|
Protein Class |
|
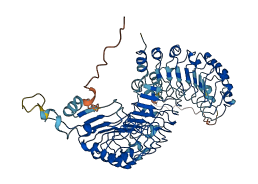
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q7SXW3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q7SXW3-F1 | Predicted | AlphaFoldDB |
No variants for Q7SXW3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q7SXW3 | |||||
No associated diseases with Q7SXW3
21 regional properties for Q7SXW3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 81 - 148 | IPR001611-1 |
| repeat | Leucine-rich repeat | 150 - 172 | IPR001611-2 |
| repeat | Leucine-rich repeat | 173 - 195 | IPR001611-3 |
| repeat | Leucine-rich repeat | 196 - 263 | IPR001611-4 |
| repeat | Leucine-rich repeat | 264 - 333 | IPR001611-5 |
| repeat | Leucine-rich repeat | 449 - 506 | IPR001611-6 |
| repeat | Leucine-rich repeat | 517 - 575 | IPR001611-7 |
| repeat | Leucine-rich repeat, typical subtype | 80 - 101 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 102 - 124 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 125 - 147 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 148 - 170 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 171 - 193 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 194 - 217 | IPR003591-6 |
| repeat | Leucine-rich repeat, typical subtype | 240 - 262 | IPR003591-7 |
| repeat | Leucine-rich repeat, typical subtype | 286 - 308 | IPR003591-8 |
| repeat | Leucine-rich repeat, typical subtype | 309 - 330 | IPR003591-9 |
| repeat | Leucine-rich repeat, typical subtype | 332 - 355 | IPR003591-10 |
| repeat | Leucine-rich repeat, typical subtype | 449 - 469 | IPR003591-11 |
| repeat | Leucine-rich repeat, typical subtype | 470 - 492 | IPR003591-12 |
| repeat | Leucine-rich repeat, typical subtype | 493 - 516 | IPR003591-13 |
| repeat | Leucine-rich repeat, typical subtype | 540 - 563 | IPR003591-14 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| protein serine/threonine phosphatase activity | Catalysis of the reaction: protein serine phosphate + H2O = protein serine + phosphate, and protein threonine phosphate + H2O = protein threonine + phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSRFKRGAHV | DSGAGFRQEK | EDCPVPYGLL | KAARKSGQLN | LSGRGLTEVP | ASVWRLNLDT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PQEAKQNVSF | GAEDRWWEQT | DLTKLLLSSN | KLQSIPDDVK | LLPALVVLDI | HDNQLSSLPD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SIGDLEQLQK | LILSHNKLTE | LPSGVWRLTN | LRCLHLQQNL | IEQIPRDLGQ | LVNLDELDLS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NNHLIDIPES | LANLQNLVKL | DLSCNKLKSL | PPAISQMKNL | RMLDCSRNQM | ESIPPVLAQM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESLEQLYLRH | NKLRYLPELP | CCKTLKELHC | GNNQIEVLEA | EHLKHLNALS | LLELRDNKVK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SLPEEITLLQ | GLERLDLTNN | DISSLPCGLG | TLPKLKSLSL | EGNPLRAIRR | DLLTKGTGEL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LKYLRSRVQE | PPNGGLKEEP | KTAMTFPSQA | KINVHAIKTL | KTLDYSEKQD | ATIPDDVFDA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VDGNPVANVN | FSKNQLTAVP | HRIVDLKDSL | ADINLGFNKL | TTIPADFCHL | KQLMHIDLRN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| NLLISLPMEL | EGLIKLRSVI | LSFNRFKSFP | EVLYRIPSLE | TILISSNQVG | GIDAVQMKTL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SRLSTLDLSN | NDIMQVPPEL | GNCTSLRALM | LDGNPFRNPR | AAILIKGTDA | VLEYLRSRIP |
| T |