Q7LBR1
Gene name |
CHMP1B (C18orf2) |
Protein name |
Charged multivesicular body protein 1b |
Names |
CHMP1.5, Chromatin-modifying protein 1b, CHMP1b, Vacuolar protein sorting-associated protein 46-2, Vps46-2, hVps46-2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:57132 |
EC number |
|
Protein Class |
|
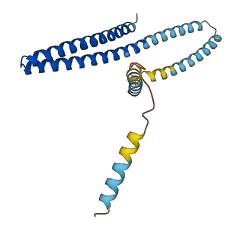
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

9 structures for Q7LBR1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3EAB | X-ray | 250 A | G/H/I/J/K/L | 148-197 | PDB |
| 3JC1 | EM | 400 A | Ab/Ad/Af/Ah/Aj/Al/An/Ap/Ar/At/Av/Ax/Az/Bb/Bd/Bf/Bh/Bj/Bl/Bn/Bp/Br/Bt/Bv/Bx/Bz/Cb/Cd/Cf/Ch | 4-163 | PDB |
| 4TXQ | X-ray | 221 A | C/D | 176-199 | PDB |
| 4TXR | X-ray | 100 A | B | 176-199 | PDB |
| 6E8G | EM | 290 A | AA/AB/B/CA/CB/D/EA/EB/F/GA/GB/H/IA/IB/J/KA/KB/L/MA/MB/N/OA/OB/P/QA/QB/R/SA/SB/T | 1-199 | PDB |
| 6TZ4 | EM | 320 A | 02/A/BA/BB/C/DA/DB/E/FA/FB/G/HA/HB/I/JA/JB/K/LA/LB/M/NA/NB/O/PA/PB/Q/RA/RB/S/TA | 1-199 | PDB |
| 6TZ5 | EM | 310 A | AA/AB/B/CA/CB/D/EA/EB/F/GA/GB/H/IA/IB/J/KA/KB/L/MA/MB/N/OA/OB/P/QA/QB/R/SA/T/UA | 1-199 | PDB |
| 6TZ9 | EM | 620 A | A/AA/B/C/D/E/F/G/H/I/J/K/L/M/N/O/P/Q/R/S/T/V/W/X/Y/Z | 1-199 | PDB |
| AF-Q7LBR1-F1 | Predicted | AlphaFoldDB |
177 variants for Q7LBR1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA296055540 rs980878125 |
3 | N>D | No |
ClinGen TOPMed gnomAD |
|
|
CA401928491 rs749979805 |
4 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA8893988 rs749979805 |
4 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs760445799 CA8893989 |
6 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8893991 CA296055553 rs376388065 |
7 | H>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8893990 rs766642864 |
7 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1237603863 CA401928520 |
8 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs755446778 CA8893992 |
9 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1360455718 CA401928527 |
9 | F>S | No |
ClinGen TOPMed |
|
|
CA8893993 rs779354092 |
13 | F>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8893995 rs753226487 |
14 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1397382348 CA401928559 |
14 | A>T | No |
ClinGen gnomAD |
|
|
CA8893994 rs753226487 |
14 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1419644692 CA401928570 |
16 | K>E | No |
ClinGen TOPMed |
|
|
CA401928572 rs777911714 |
16 | K>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1259545003 CA401928575 |
16 | K>N | No |
ClinGen TOPMed |
|
|
rs777911714 CA8893996 |
16 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747190624 CA8893997 |
17 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA401928585 rs1188161321 |
18 | L>R | No |
ClinGen TOPMed |
|
|
CA401928590 rs1486157193 |
19 | S>C | No |
ClinGen TOPMed |
|
|
rs771241756 CA8893998 |
19 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1211964016 CA401928604 |
21 | S>G | No |
ClinGen TOPMed |
|
| TCGA novel | 24 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756274240 | 25 | C>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs749131244 CA8894001 |
25 | C>W | No |
ClinGen ExAC gnomAD |
|
|
rs1232508978 CA401928635 |
25 | C>Y | No |
ClinGen gnomAD |
|
|
CA8894002 rs768600593 |
26 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 26 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA401928647 rs1216651201 |
27 | K>E | No |
ClinGen gnomAD |
|
|
CA401928649 rs1257524312 |
27 | K>T | No |
ClinGen gnomAD |
|
|
CA401928660 rs1567892083 |
28 | E>D | No |
ClinGen Ensembl |
|
|
rs368781162 CA8894004 |
28 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA401928674 rs1298225930 |
30 | K>N | No |
ClinGen TOPMed |
|
|
CA296055580 rs1053039371 |
30 | K>R | No |
ClinGen gnomAD |
|
|
CA8894007 rs555902532 |
31 | A>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA401928680 rs1189119978 |
31 | A>V | No |
ClinGen gnomAD |
|
|
CA296055587 rs779086835 |
32 | E>G | No |
ClinGen Ensembl |
|
|
CA296055605 rs933590082 |
34 | A>V | No |
ClinGen Ensembl |
|
|
rs760223704 CA8894009 |
35 | K>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 36 | I>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA401928729 rs1374126927 |
38 | K>N | No |
ClinGen gnomAD |
|
|
rs1462108140 CA401928731 |
39 | A>T | No |
ClinGen gnomAD |
|
|
rs1264189524 CA401928736 |
39 | A>V | No |
ClinGen gnomAD |
|
|
CA401928738 rs1383199965 |
40 | I>V | No |
ClinGen TOPMed |
|
|
rs1567892135 CA401928762 |
43 | G>S | No |
ClinGen Ensembl |
|
|
COSM1732947 CA8894012 rs759848382 |
44 | N>S | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA401928769 rs759848382 |
44 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8894013 rs577828370 |
45 | M>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA8894014 rs577828370 |
45 | M>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs758967526 CA8894015 |
48 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758967526 CA401928797 |
48 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8894016 rs372758165 |
49 | R>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372758165 CA296055625 |
49 | R>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1461190700 CA401928812 |
51 | H>D | No |
ClinGen TOPMed |
|
|
CA401928817 rs1371062795 |
51 | H>Q | No |
ClinGen TOPMed |
|
|
rs1294858936 CA401928837 |
54 | N>S | No |
ClinGen gnomAD |
|
|
CA8894019 rs757390250 |
57 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA296055641 rs1050860397 |
57 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1260068235 CA401928862 |
58 | Q>R | No |
ClinGen gnomAD |
|
|
CA8894021 rs200810906 |
59 | K>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs768510996 CA8894022 |
60 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA401928876 rs1193387832 |
60 | N>T | No |
ClinGen gnomAD |
|
|
rs1007245929 CA296055647 |
61 | Q>* | No |
ClinGen TOPMed |
|
|
CA8894023 rs201261255 |
62 | A>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs201261255 CA401928890 |
62 | A>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA401928896 rs1598428156 |
63 | V>G | No |
ClinGen Ensembl |
|
|
rs1171945498 CA401928906 |
65 | F>I | No |
ClinGen gnomAD |
|
|
CA401928940 rs1253384854 |
69 | S>N | No |
ClinGen TOPMed |
|
|
rs1376895764 CA401928945 |
70 | A>T | No |
ClinGen gnomAD |
|
|
CA8894026 rs772566478 |
71 | R>P | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 72 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1266710923 CA401928966 |
73 | D>V | No |
ClinGen gnomAD |
|
|
rs760196342 CA8894028 |
77 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA8894031 rs776210771 |
78 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs765579637 CA8894032 |
79 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1301933761 CA401929010 |
80 | Q>H | No |
ClinGen TOPMed |
|
|
rs993999075 CA296055672 |
83 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA8894033 rs775888398 |
84 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA8894034 rs763343480 |
86 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA401929051 rs372085953 |
87 | K>M | No |
ClinGen ESP ExAC gnomAD |
|
|
rs372085953 CA8894035 |
87 | K>R | No |
ClinGen ESP ExAC gnomAD |
|
|
rs529426406 CA8894036 |
88 | V>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1422738305 CA401929070 |
90 | K>T | No |
ClinGen TOPMed gnomAD |
|
|
CA8894037 rs757310923 |
92 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA401929092 rs952579364 |
93 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
rs952579364 CA296055698 |
93 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs767506161 CA8894038 |
94 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA401929122 rs1193683005 |
98 | S>W | No |
ClinGen gnomAD |
|
|
CA8894041 rs188014726 |
102 | T>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA401929149 rs1158139986 |
102 | T>S | No |
ClinGen gnomAD |
|
|
rs747997562 CA8894042 |
106 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1291773728 CA401929174 |
106 | M>V | No |
ClinGen gnomAD |
|
| TCGA novel | 109 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8894044 rs777720728 |
111 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs1378206044 CA401929227 |
113 | A>V | No |
ClinGen TOPMed |
|
| TCGA novel | 114 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1374997744 CA401929241 |
115 | M>I | No |
ClinGen gnomAD |
|
|
rs1567892310 CA401929235 |
115 | M>V | No |
ClinGen Ensembl |
|
|
CA401929249 rs1223367435 |
116 | D>G | No |
ClinGen gnomAD |
|
|
rs770409909 CA8894046 |
117 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA401929281 rs776188158 |
119 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA8894047 rs776188158 |
119 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1436274813 CA401929312 |
121 | Q>H | No |
ClinGen TOPMed |
|
|
CA401929317 rs1273156911 |
122 | F>L | No |
ClinGen gnomAD |
|
|
rs1469289774 CA401929372 |
127 | V>A | No |
ClinGen gnomAD |
|
|
rs1456992668 CA401929386 |
128 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1485576961 CA401929409 |
130 | Q>L | No |
ClinGen TOPMed |
|
|
rs745503644 CA8894048 |
131 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs769482223 CA8894049 |
131 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA296055752 rs981760833 |
132 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA8894050 rs775796754 |
132 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA8894051 rs763336856 |
135 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA8894052 rs764387927 |
135 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs562746140 CA8894054 |
138 | S>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA401929517 rs1328011169 |
139 | T>A | No |
ClinGen gnomAD |
|
|
rs377323477 CA401929546 |
142 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8894056 rs377323477 |
142 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1366864125 CA401929558 |
143 | T>S | No |
ClinGen gnomAD |
|
|
CA8894059 rs754140300 |
144 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA401929566 rs1407372558 |
144 | T>N | No |
ClinGen gnomAD |
|
|
CA8894060 rs758158028 CA401929617 |
148 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1340313610 CA401929636 |
150 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs551779348 CA8894062 CA8894063 |
151 | M>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1374586589 CA401929644 |
151 | M>V | No |
ClinGen TOPMed |
|
|
CA401929660 rs1310853841 |
152 | L>R | No |
ClinGen TOPMed |
|
|
CA401929655 rs1435626999 |
152 | L>V | No |
ClinGen gnomAD |
|
|
rs915372954 CA296055794 |
155 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs779631894 CA8894067 |
155 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA401929728 rs1308250707 |
158 | D>E | No |
ClinGen gnomAD |
|
| TCGA novel | 159 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA401929744 rs1221080522 |
160 | A>T | No |
ClinGen gnomAD |
|
|
rs1256462232 CA401929758 |
161 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs748904850 CA8894068 |
162 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1598428477 CA401929777 |
163 | D>A | No |
ClinGen Ensembl |
|
| TCGA novel | 163 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA401929783 rs1289732853 |
164 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA8894070 rs774638280 |
165 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA8894071 rs774638280 |
165 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA8894072 rs772746987 |
166 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA401929810 rs1598428505 |
166 | M>R | No |
ClinGen Ensembl |
|
|
CA401929805 rs772746987 |
166 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773886733 CA8894073 |
167 | E>* | No |
ClinGen ExAC |
|
|
rs760793456 CA296055805 |
167 | E>A | No |
ClinGen ExAC |
|
|
CA8894074 rs760793456 |
167 | E>V | No |
ClinGen ExAC |
|
|
CA401929836 rs1356518735 |
169 | P>S | No |
ClinGen gnomAD |
|
|
CA8894077 rs534105520 |
171 | G>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA401929857 rs1312654764 |
171 | G>S | No |
ClinGen gnomAD |
|
|
CA8894079 rs759746944 CA401929874 |
172 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763933033 CA8894080 |
173 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA401929891 rs1262011494 |
174 | G>A | No |
ClinGen TOPMed |
|
|
CA401929887 rs1213528655 |
174 | G>R | No |
ClinGen gnomAD |
|
|
CA8894082 rs757150985 |
175 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA8894084 rs549187606 |
176 | V>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs549187606 CA8894085 |
176 | V>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA401929920 rs1485915465 |
177 | G>D | No |
ClinGen gnomAD |
|
|
CA401929916 rs1262309666 |
177 | G>R | No |
ClinGen gnomAD |
|
|
rs1267206062 CA401929927 |
178 | T>K | No |
ClinGen Ensembl |
|
|
CA401929942 rs1240339384 |
180 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1161280273 CA401929958 |
181 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
rs145696105 CA8894089 |
181 | A>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs748422423 CA8894090 |
182 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772587665 CA401929974 |
183 | A>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs772587665 CA401929975 |
183 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772587665 CA8894091 |
183 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1376283177 CA401929981 |
184 | E>K | No |
ClinGen gnomAD |
|
|
CA8894093 rs761272544 |
186 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA401930007 rs761272544 |
186 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1296713863 CA401930028 |
188 | L>V | No |
ClinGen gnomAD |
|
|
CA401930040 rs1270286694 |
189 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
CA8894095 rs771133779 |
189 | S>P | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 191 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs759779413 CA8894097 |
194 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs765415090 CA8894098 |
195 | L>F | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 196 | R>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA401930112 rs1473358823 |
196 | R>L | No |
ClinGen TOPMed |
|
|
rs1191620408 CA401930125 |
198 | Q>K | No |
ClinGen TOPMed |
|
|
rs761542966 CA8894100 |
199 | V>M | No |
ClinGen ExAC gnomAD |
No associated diseases with Q7LBR1
No regional properties for Q7LBR1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q7LBR1 | |||
Functions
16 GO annotations of cellular component
| Name | Definition |
|---|---|
| amphisome membrane | Any membrane that is part of an amphisome. |
| autophagosome membrane | The lipid bilayer surrounding an autophagosome, a double-membrane-bounded vesicle in which endogenous cellular material is sequestered. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| ESCRT III complex | A complex with membrane scission activity that plays a major role in many processes where membranes are remodelled - including endosomal transport (vesicle budding), nuclear envelope organisation (membrane closure, mitotic bridge cleavage), and cytokinesis (abscission). |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| kinetochore | A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules. |
| kinetochore microtubule | Any of the spindle microtubules that attach to the kinetochores of chromosomes by their plus ends, and maneuver the chromosomes during mitotic or meiotic chromosome segregation. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| membrane coat | Any of several different proteinaceous coats that can associate with membranes. Membrane coats include those formed by clathrin plus an adaptor complex, the COPI and COPII complexes, and possibly others. They are found associated with membranes on many vesicles as well as other membrane features such as pits and perhaps tubules. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| multivesicular body | A type of endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body membrane | The lipid bilayer surrounding a multivesicular body. |
| nuclear pore | A protein complex providing a discrete opening in the nuclear envelope of a eukaryotic cell, where the inner and outer nuclear membranes are joined. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| identical protein binding | Binding to an identical protein or proteins. |
| MIT domain binding | Binding to a MIT protein domain. The MIT domain is found in vacuolar sorting proteins, spastin (probable ATPase involved in the assembly or function of nuclear protein complexes), and a sorting nexin, which may play a role in intracellular trafficking. |
| protein domain specific binding | Binding to a specific domain of a protein. |
25 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome maturation | Removal of PI3P and Atg8/LC3 after the closure of the phagophore and before the fusion with the endosome/lysosome (e.g. mammals and insects) or vacuole (yeast), and that very likely destabilizes other Atg proteins and thus enables their efficient dissociation and recycling. |
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| endosome transport via multivesicular body sorting pathway | The directed movement of substances from endosomes to lysosomes or vacuoles by a pathway in which molecules are sorted into multivesicular bodies, which then fuse with the target compartment. |
| ESCRT III complex disassembly | The disaggregation of an ESCRT III complex into its constituent components. |
| establishment of protein localization | The directed movement of a protein to a specific location. |
| late endosome to lysosome transport | The directed movement of substances from late endosome to lysosome. |
| late endosome to vacuole transport | The directed movement of substances from late endosomes to the vacuole. In yeast, after transport to the prevacuolar compartment, endocytic content is delivered to the late endosome and on to the vacuole. This pathway is analogous to endosome to lysosome transport. |
| membrane fission | A process that is carried out at the cellular level which results in the separation of a single continuous membrane into two membranes. |
| midbody abscission | The process by which the midbody, the cytoplasmic bridge that connects the two prospective daughter cells, is severed at the end of mitotic cytokinesis, resulting in two separate daughter cells. |
| mitotic metaphase plate congression | The cell cycle process in which chromosomes are aligned at the metaphase plate, a plane halfway between the poles of the mitotic spindle, during mitosis. |
| multivesicular body assembly | The aggregation, arrangement and bonding together of a set of components to form a multivesicular body, a type of late endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body sorting pathway | A vesicle-mediated transport process in which transmembrane proteins are ubiquitylated to facilitate their entry into luminal vesicles of multivesicular bodies (MVBs); upon subsequent fusion of MVBs with lysosomes or vacuoles, the cargo proteins are degraded. |
| multivesicular body-lysosome fusion | The organelle membrane fusion process in which the membrane of a multivesicular body fuses with a lysosome to create a hybrid organelle. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| nuclear membrane reassembly | The reformation of the nuclear membranes following their breakdown in the context of a normal process. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| plasma membrane repair | The resealing of a cell plasma membrane after cellular wounding due to, for instance, mechanical stress. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of centrosome duplication | Any process that modulates the frequency, rate or extent of centrosome duplication. Centrosome duplication is the replication of a centrosome, a structure comprised of a pair of centrioles and peri-centriolar material from which a microtubule spindle apparatus is organized. |
| regulation of mitotic spindle assembly | Any process that modulates the frequency, rate or extent of mitotic spindle assembly. |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide covalently tagged with ubiquitin, via the multivesicular body (MVB) sorting pathway; ubiquitin-tagged proteins are sorted into MVBs, and delivered to a lysosome/vacuole for degradation. |
| vesicle fusion with vacuole | The joining of the lipid bilayer membrane around a vesicle with the lipid bilayer membrane around the vacuole. |
| viral budding from plasma membrane | A viral budding that starts with formation of a membrane curvature in the host plasma membrane. |
| viral budding via host ESCRT complex | Viral budding which uses a host ESCRT protein complex, or complexes, to mediate the budding process. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E994 | CHMP1B | Charged multivesicular body protein 1b | Bos taurus (Bovine) | PR |
| Q5ZKX1 | CHMP1B | Charged multivesicular body protein 1b | Gallus gallus (Chicken) | PR |
| O43633 | CHMP2A | Charged multivesicular body protein 2a | Homo sapiens (Human) | EV |
| Q9Y3E7 | CHMP3 | Charged multivesicular body protein 3 | Homo sapiens (Human) | EV |
| Q9CQD4 | Chmp1b2 | Charged multivesicular body protein 1b-2 | Mus musculus (Mouse) | PR |
| Q99LU0 | Chmp1b1 | Charged multivesicular body protein 1b-1 | Mus musculus (Mouse) | PR |
| Q84VG1 | CHMP1 | ESCRT-related protein CHMP1 | Oryza sativa subsp japonica (Rice) | PR |
| Q8LE58 | CHMP1A | ESCRT-related protein CHMP1A | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSM4 | CHMP1B | ESCRT-related protein CHMP1B | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6DF27 | chmp1b | Charged multivesicular body protein 1b | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q7ZVB1 | chmp1b | Charged multivesicular body protein 1b | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSNMEKHLFN | LKFAAKELSR | SAKKCDKEEK | AEKAKIKKAI | QKGNMEVARI | HAENAIRQKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QAVNFLRMSA | RVDAVAARVQ | TAVTMGKVTK | SMAGVVKSMD | ATLKTMNLEK | ISALMDKFEH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QFETLDVQTQ | QMEDTMSSTT | TLTTPQNQVD | MLLQEMADEA | GLDLNMELPQ | GQTGSVGTSV |
| 190 | |||||
| ASAEQDELSQ | RLARLRDQV |