Q7L9L4
Gene name |
MOB1B (MOB4A, MOBKL1A) |
Protein name |
MOB kinase activator 1B |
Names |
Mob1 homolog 1A , Mob1A , Mob1B , Mps one binder kinase activator-like 1A |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:92597 |
EC number |
|
Protein Class |
|
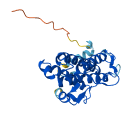
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
41-204 (MH domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q7L9L4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q7L9L4-F1 | Predicted | AlphaFoldDB |
210 variants for Q7L9L4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1304822460 | 2 | S>N | No |
TOPMed gnomAD |
|
| rs1258096363 | 3 | F>I | No | gnomAD | |
| rs1314098870 | 3 | F>L | No |
TOPMed gnomAD |
|
| rs1357225726 | 4 | L>* | No |
TOPMed gnomAD |
|
| rs757952906 | 4 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1357225726 | 4 | L>S | No |
TOPMed gnomAD |
|
| rs1237174047 | 5 | F>I | No |
TOPMed gnomAD |
|
| rs1237174047 | 5 | F>L | No |
TOPMed gnomAD |
|
| rs777510058 | 5 | F>S | No |
ExAC gnomAD |
|
| rs1237174047 | 5 | F>V | No |
TOPMed gnomAD |
|
| rs776857746 | 7 | S>G | No |
ExAC gnomAD |
|
| rs759473680 | 7 | S>R | No |
ExAC gnomAD |
|
| rs769925167 | 8 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs775501483 | 8 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs763506781 | 9 | S>F | No |
ExAC gnomAD |
|
| rs764750751 | 10 | S>F | No |
ExAC gnomAD |
|
| rs1179125921 | 11 | K>N | No | gnomAD | |
| COSM1057148 | 11 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 12 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751942946 | 13 | F>C | No |
ExAC gnomAD |
|
| rs751942946 | 13 | F>S | No |
ExAC gnomAD |
|
| TCGA novel | 13 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1230887637 | 14 | K>T | No | gnomAD | |
| rs762300488 | 15 | P>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 16 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1738182090 | 16 | K>T | No | gnomAD | |
| rs767231417 | 17 | K>E | No |
ExAC gnomAD |
|
| rs750105577 | 17 | K>M | No |
ExAC gnomAD |
|
| rs750105577 | 17 | K>R | No |
ExAC gnomAD |
|
| rs755788328 | 18 | N>S | No |
ExAC gnomAD |
|
| rs1480418127 | 19 | I>F | No |
TOPMed gnomAD |
|
| rs1480418127 | 19 | I>V | No |
TOPMed gnomAD |
|
| rs139701003 | 20 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs753337742 | 21 | E>A | No |
ExAC gnomAD |
|
| rs1738183512 | 22 | G>V | No | TOPMed | |
| rs1578388989 | 23 | S>F | No | TOPMed | |
| rs754877537 | 23 | S>P | No | ExAC | |
| COSM3775959 | 24 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs961340952 | 25 | Q>R | No | Ensembl | |
| rs1738184143 | 26 | Y>N | No | TOPMed | |
| rs1738184257 | 27 | E>K | No |
TOPMed gnomAD |
|
| rs1738184366 | 28 | L>P | No | Ensembl | |
| rs1578388997 | 29 | L>* | No | Ensembl | |
| rs972778706 | 31 | H>Y | No | Ensembl | |
| rs1578389026 | 32 | A>E | No | Ensembl | |
| rs747885745 | 32 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM1430821 rs747885745 |
32 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs771865876 | 33 | E>K | No |
ExAC gnomAD |
|
| rs1479110287 | 34 | A>D | No | gnomAD | |
| rs1738186353 | 38 | S>C | No | Ensembl | |
| rs1355471384 | 39 | G>D | No |
TOPMed gnomAD |
|
| rs1461137358 | 40 | N>K | No | gnomAD | |
| rs746102820 | 40 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs746102820 | 40 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs769971173 | 41 | L>F | No |
ExAC gnomAD |
|
| rs775621553 | 42 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM1540916 rs1392541792 COSM6168141 |
42 | R>W | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1302388497 | 43 | M>I | No | gnomAD | |
| rs1738187483 | 44 | A>T | No |
TOPMed gnomAD |
|
| rs1738187715 | 45 | V>G | No | TOPMed | |
| rs1307028363 | 46 | M>I | No | gnomAD | |
| rs868771099 | 46 | M>T | No | Ensembl | |
| rs762987346 | 46 | M>V | No |
ExAC gnomAD |
|
| rs144398833 | 48 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs775021039 | 49 | E>K | No |
ExAC gnomAD |
|
| rs1738188820 | 50 | G>A | No | Ensembl | |
| rs762212825 | 50 | G>R | No |
ExAC gnomAD |
|
| rs1362713671 | 51 | E>D | No |
TOPMed gnomAD |
|
| rs200143951 | 52 | D>N | No |
1000Genomes ExAC gnomAD |
|
| rs1264404395 | 52 | D>V | No | gnomAD | |
| TCGA novel | 52 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146589415 | 54 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1424440858 | 56 | W>* | No |
TOPMed gnomAD |
|
| rs1578389163 | 57 | V>F | No | Ensembl | |
| rs757025470 | 62 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1459599024 | 66 | N>S | No | gnomAD | |
| rs1738690793 | 69 | N>D | No | TOPMed | |
| rs1441239173 | 70 | M>I | No |
TOPMed gnomAD |
|
| rs1254186698 | 70 | M>K | No |
TOPMed gnomAD |
|
| rs755320738 | 70 | M>L | No |
ExAC gnomAD |
|
| rs755320738 | 70 | M>V | No |
ExAC gnomAD |
|
| rs1485546616 | 74 | T>S | No | TOPMed | |
| rs748277810 | 75 | I>V | No |
ExAC gnomAD |
|
| rs772591968 | 76 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs532138438 | 76 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs2148899751 | 78 | F>V | No | Ensembl | |
| rs1332205033 | 79 | C>R | No | gnomAD | |
| rs376624270 | 80 | T>I | No |
ESP TOPMed gnomAD |
|
| rs1329296490 | 82 | E>K | No |
TOPMed gnomAD |
|
| rs759178033 | 83 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1412619524 | 84 | C>G | No | TOPMed | |
| rs1034163960 | 84 | C>Y | No |
TOPMed gnomAD |
|
| rs1560662795 | 85 | P>A | No | gnomAD | |
| rs1560662795 | 85 | P>S | No | gnomAD | |
| rs1738694340 | 87 | M>I | No | TOPMed | |
| rs1334501500 | 87 | M>K | No |
TOPMed gnomAD |
|
| rs1560666024 | 93 | Y>H | No | Ensembl | |
| rs372659983 | 94 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs761645813 | 96 | H>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 97 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1276964796 | 98 | A>T | No |
TOPMed gnomAD |
|
| rs1414797226 | 98 | A>V | No | gnomAD | |
| rs767528335 | 101 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs767528335 | 101 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs760568838 | 102 | N>S | No |
ExAC gnomAD |
|
| rs1738933154 | 103 | I>V | No | TOPMed | |
| TCGA novel | 104 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765493444 | 104 | K>R | No |
ExAC gnomAD |
|
| rs767123707 | 105 | K>N | No |
TOPMed gnomAD |
|
| rs758710829 | 107 | I>S | No |
ExAC gnomAD |
|
| rs752969116 | 107 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1247747287 | 110 | S>F | No | gnomAD | |
| rs777699597 | 110 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs938145855 | 111 | A>G | No | TOPMed | |
| rs377705076 | 111 | A>S | No |
ESP ExAC gnomAD |
|
|
COSM195617 rs377705076 |
111 | A>T | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC gnomAD |
| rs781689627 | 113 | K>E | No |
ExAC TOPMed gnomAD |
|
|
rs1286249802 COSM448098 |
114 | Y>C | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| COSM3605546 | 115 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747038359 | 117 | Y>* | No |
ExAC TOPMed gnomAD |
|
| COSM1328417 | 119 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1738935762 | 119 | M>V | No | Ensembl | |
| rs779615480 | 120 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1738936101 | 121 | W>C | No |
TOPMed gnomAD |
|
| rs1738936265 | 123 | Q>* | No | TOPMed | |
| TCGA novel | 124 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1238176695 | 124 | D>H | No | gnomAD | |
| rs1446624388 | 125 | Q>E | No | gnomAD | |
| rs140427584 | 130 | T>M | No |
ESP TOPMed gnomAD |
|
| rs140427584 | 130 | T>R | No |
ESP TOPMed gnomAD |
|
| rs987127183 | 131 | L>S | No | Ensembl | |
| rs912374825 | 134 | S>P | No | Ensembl | |
| rs1160893401 | 135 | K>I | No | gnomAD | |
| COSM1057150 | 136 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1738937786 | 136 | I>V | No | TOPMed | |
| rs777892134 | 139 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1197808536 | 140 | F>L | No |
TOPMed gnomAD |
|
| rs199526060 | 140 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs1197808536 | 140 | F>V | No |
TOPMed gnomAD |
|
| rs370951801 | 142 | K>E | No |
ESP TOPMed gnomAD |
|
| COSM5369462 | 142 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs537448770 | 144 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs764175274 | 144 | F>Y | No |
ExAC TOPMed gnomAD |
|
| rs1434267276 | 145 | M>I | No |
TOPMed gnomAD |
|
| rs1295196250 | 145 | M>V | No | TOPMed | |
| rs201445824 | 147 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1479767686 | 148 | A>P | No | gnomAD | |
| rs1479767686 | 148 | A>S | No | gnomAD | |
| rs1456406840 | 150 | T>S | No | TOPMed | |
| rs750563749 | 151 | I>M | No |
ExAC gnomAD |
|
| rs1047530845 | 151 | I>V | No | TOPMed | |
| rs2148904492 | 152 | L>F | No | Ensembl | |
| rs1462145428 | 153 | K>N | No |
TOPMed gnomAD |
|
| rs368157226 | 154 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
rs755380499 COSM1671064 |
154 | R>H | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs755380499 | 154 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs368157226 | 154 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1739109845 | 155 | L>F | No | TOPMed | |
| rs1739109845 | 155 | L>I | No | TOPMed | |
| rs201391108 | 156 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs771249425 | 161 | H>Q | No | Ensembl | |
| rs758181496 | 162 | I>V | No | ExAC | |
| rs370279470 | 163 | Y>C | No |
ESP TOPMed gnomAD |
|
| rs1403051348 | 165 | Q>E | No | gnomAD | |
| rs1282680304 | 166 | H>R | No | gnomAD | |
| rs1739110877 | 168 | D>E | No | TOPMed | |
| rs777398554 | 169 | P>T | No |
ExAC gnomAD |
|
| rs934927817 | 171 | I>V | No | TOPMed | |
| rs1739111541 | 175 | E>* | No | Ensembl | |
| COSM6101039 | 176 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1210780641 | 179 | L>P | No | gnomAD | |
| COSM5328232 | 179 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770953916 | 181 | T>I | No |
ExAC gnomAD |
|
| rs2148904544 | 182 | S>F | No | Ensembl | |
| rs745592131 | 182 | S>T | No |
ExAC gnomAD |
|
| rs1267478332 | 183 | F>L | No |
TOPMed gnomAD |
|
| rs1428577624 | 184 | K>R | No | gnomAD | |
| rs1739112543 | 185 | H>Q | No | gnomAD | |
| rs1194779732 | 186 | F>C | No | gnomAD | |
| TCGA novel | 189 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs778232663 COSM1430822 |
189 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| COSM4615348 | 190 | V>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs908851132 | 190 | V>I | No | Ensembl | |
| rs769682033 | 194 | N>S | No |
ExAC gnomAD |
|
| rs1739226252 | 195 | L>F | No | Ensembl | |
| rs779880416 | 196 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs749124116 | 196 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs779880416 | 196 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs187615532 | 198 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs773434094 | 199 | R>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 199 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 200 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760678125 | 200 | E>K | No |
ExAC gnomAD |
|
| rs776632688 | 201 | L>F | No |
ExAC gnomAD |
|
| rs760065451 | 202 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs1338822244 | 202 | A>T | No | gnomAD | |
| rs1739228189 | 203 | P>S | No | TOPMed | |
| rs372253364 | 205 | Q>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1739228539 | 206 | E>* | No | TOPMed | |
| rs1256450687 | 207 | L>M | No |
TOPMed gnomAD |
|
| rs1222508125 | 207 | L>P | No | gnomAD | |
| TCGA novel | 207 | L>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 208 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751262553 | 209 | E>G | No |
ExAC gnomAD |
|
| rs1344217508 | 210 | K>E | No | gnomAD | |
| rs1739229801 | 212 | T>P | No | Ensembl | |
| rs780744296 | 213 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs780744296 | 213 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs865915578 | 215 | D>G | No | Ensembl | |
| rs779889508 | 217 | R>Y | No |
ExAC TOPMed gnomAD |
No associated diseases with Q7L9L4
No regional properties for Q7L9L4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q7L9L4 | |||
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| kinase activator activity | Binds to and increases the activity of a kinase, an enzyme which catalyzes of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| kinase binding | Binding to a kinase, any enzyme that catalyzes the transfer of a phosphate group. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activator activity | Binds to and increases the activity of a protein kinase, an enzyme which phosphorylates a protein. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| hippo signaling | The series of molecular signals mediated by the serine/threonine kinase Hippo or one of its orthologs. In Drosophila, Hippo in complex with the scaffold protein Salvador (Sav), phosphorylates and activates Warts (Wts), which in turn phosphorylates and inactivates the Yorkie (Yki) transcriptional activator. The core fly components hippo, sav, wts and mats are conserved in mammals as STK4/3 (MST1/2), SAV1/WW45, LATS1/2 and MOB1. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| regulation of protein autophosphorylation | Any process that modulates the frequency, rate or extent of addition of the phosphorylation by a protein of one or more of its own residues. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P43563 | MOB2 | CBK1 kinase activator protein MOB2 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P40484 | MOB1 | DBF2 kinase activator protein MOB1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| Q95RA8 | mats | MOB kinase activator-like 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q70IA8 | MOB3C | MOB kinase activator 3C | Homo sapiens (Human) | SS |
| Q96BX8 | MOB3A | MOB kinase activator 3A | Homo sapiens (Human) | SS |
| Q86TA1 | MOB3B | MOB kinase activator 3B | Homo sapiens (Human) | SS |
| Q9H8S9 | MOB1A | MOB kinase activator 1A | Homo sapiens (Human) | EV |
| Q921Y0 | Mob1a | MOB kinase activator 1A | Mus musculus (Mouse) | SS |
| Q8VI63 | Mob2 | MOB kinase activator 2 | Mus musculus (Mouse) | EV |
| Q8BPB0 | Mob1b | MOB kinase activator 1B | Mus musculus (Mouse) | EV |
| Q3T1J9 | Mob1a | MOB kinase activator 1A | Rattus norvegicus (Rat) | SS |
| Q8GYX0 | MOB1B | MOB kinase activator-like 1B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHI1 | MOB1A | MOB kinase activator-like 1A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSFLFGSRSS | KTFKPKKNIP | EGSHQYELLK | HAEATLGSGN | LRMAVMLPEG | EDLNEWVAVN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TVDFFNQINM | LYGTITDFCT | EESCPVMSAG | PKYEYHWADG | TNIKKPIKCS | APKYIDYLMT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| WVQDQLDDET | LFPSKIGVPF | PKNFMSVAKT | ILKRLFRVYA | HIYHQHFDPV | IQLQEEAHLN |
| 190 | 200 | 210 | |||
| TSFKHFIFFV | QEFNLIDRRE | LAPLQELIEK | LTSKDR |