Q7DMA9
Gene name |
PAS1 (DEI1, FKBP70, FKBP72, At3g54010, F5K20.310) |
Protein name |
Peptidyl-prolyl cis-trans isomerase PASTICCINO1 |
Names |
70 kDa peptidyl-prolyl isomerase, FK506-binding protein 72, AtFKBP72, Immunophilin FKBP72, Peptidyl-prolyl cis-trans isomerase FKBP72, PPIase FKBP72, Rotamase |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G54010 |
EC number |
5.2.1.8: Cis-trans isomerases |
Protein Class |
|
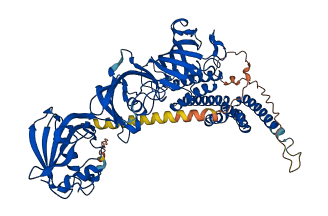
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q7DMA9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q7DMA9-F1 | Predicted | AlphaFoldDB |
48 variants for Q7DMA9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_3_20001079_C_T | 13 | P>L | No | 1000Genomes | |
| ENSVATH12751207 | 23 | D>E | No | 1000Genomes | |
| ENSVATH06311425 | 38 | V>L | No | 1000Genomes | |
| ENSVATH06311427 | 47 | S>G | No | 1000Genomes | |
| tmp_3_20001294_G_T | 47 | S>I | No | 1000Genomes | |
| tmp_3_20001396_T_C | 58 | C>R | No | 1000Genomes | |
| ENSVATH06311431 | 65 | G>A | No | 1000Genomes | |
| tmp_3_20001440_G_C | 72 | R>S | No | 1000Genomes | |
| ENSVATH06311432 | 75 | S>C | No | 1000Genomes | |
| ENSVATH06311435 | 81 | P>Q | No | 1000Genomes | |
| ENSVATH12751214 | 84 | D>G | No | 1000Genomes | |
| ENSVATH00418377 | 88 | N>K | No | 1000Genomes | |
| tmp_3_20001624_C_A | 103 | H>N | No | 1000Genomes | |
| ENSVATH00418379 | 131 | F>V | No | 1000Genomes | |
| tmp_3_20001940_T_A | 138 | H>Q | No | 1000Genomes | |
| ENSVATH06311441 | 145 | D>E | No | 1000Genomes | |
| tmp_3_20002240_A_T | 165 | E>D | No | 1000Genomes | |
| tmp_3_20002284_C_G | 180 | A>G | No | 1000Genomes | |
| tmp_3_20002452_C_G | 199 | P>A | No | 1000Genomes | |
| ENSVATH12751230 | 207 | S>Y | No | 1000Genomes | |
| ENSVATH12751231 | 218 | G>E | No | 1000Genomes | |
| ENSVATH02502316 | 239 | P>S | No | 1000Genomes | |
| tmp_3_20002674_T_C | 241 | L>S | No | 1000Genomes | |
| ENSVATH14431381 | 277 | R>Q | No | 1000Genomes | |
| ENSVATH06311450 | 287 | C>S | No | 1000Genomes | |
| tmp_3_20003071_C_T | 288 | P>L | No | 1000Genomes | |
| tmp_3_20003167_A_G | 320 | Q>R | No | 1000Genomes | |
| ENSVATH06311454 | 344 | P>T | No | 1000Genomes | |
| ENSVATH06311455 | 355 | D>G | No | 1000Genomes | |
| tmp_3_20003518_G_T | 370 | G>C | No | 1000Genomes | |
| tmp_3_20003525_A_G | 372 | H>R | No | 1000Genomes | |
| tmp_3_20003556_C_G | 382 | F>L | No | 1000Genomes | |
| ENSVATH06311460 | 399 | E>K | No | 1000Genomes | |
| tmp_3_20004064_G_A | 446 | D>N | No | 1000Genomes | |
| ENSVATH02502323 | 463 | G>E | No | 1000Genomes | |
| ENSVATH06311466 | 478 | A>G | No | 1000Genomes | |
| ENSVATH06311467 | 494 | I>N | No | 1000Genomes | |
| ENSVATH06311468 | 496 | G>A | No | 1000Genomes | |
| ENSVATH12751273 | 507 | N>D | No | 1000Genomes | |
| tmp_3_20004551_A_G | 511 | K>E | No | 1000Genomes | |
| tmp_3_20004567_C_T | 516 | S>L | No | 1000Genomes | |
| ENSVATH12751276 | 557 | G>D | No | 1000Genomes | |
| tmp_3_20004855_TAGA_GAGA,T | 567 | I>R | No | 1000Genomes | |
| ENSVATH12751277 | 573 | T>A | No | 1000Genomes | |
| ENSVATH06311477 | 585 | E>V | No | 1000Genomes | |
| ENSVATH12751278 | 603 | W>G | No | 1000Genomes | |
| ENSVATH14431384 | 605 | F>L | No | 1000Genomes | |
| ENSVATH00418387 | 618 | G>A | No | 1000Genomes |
No associated diseases with Q7DMA9
6 regional properties for Q7DMA9
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FKBP-type peptidyl-prolyl cis-trans isomerase domain | 46 - 147 | IPR001179-1 |
| domain | FKBP-type peptidyl-prolyl cis-trans isomerase domain | 175 - 256 | IPR001179-2 |
| domain | FKBP-type peptidyl-prolyl cis-trans isomerase domain | 291 - 383 | IPR001179-3 |
| repeat | Tetratricopeptide repeat | 400 - 433 | IPR019734-1 |
| repeat | Tetratricopeptide repeat | 449 - 482 | IPR019734-2 |
| repeat | Tetratricopeptide repeat | 483 - 516 | IPR019734-3 |
Functions
| Description | ||
|---|---|---|
| EC Number | 5.2.1.8 | Cis-trans isomerases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| peptidyl-prolyl cis-trans isomerase activity | Catalysis of the reaction: peptidyl-proline (omega=180) = peptidyl-proline (omega=0). |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| auxin-activated signaling pathway | The series of molecular signals generated by the binding of the plant hormone auxin to a receptor, and ending with modulation of a downstream cellular process, e.g. transcription. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| chaperone-mediated protein folding | The process of inhibiting aggregation and assisting in the covalent and noncovalent assembly of single chain polypeptides or multisubunit complexes into the correct tertiary structure that is dependent on interaction with a chaperone. |
| cytokinin-activated signaling pathway | The series of molecular signals generated by the binding of a cytokinin to a receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| embryo development ending in seed dormancy | The process whose specific outcome is the progression of the embryo over time, from zygote formation to the end of seed dormancy. An example of this process is found in Arabidopsis thaliana. |
| embryonic pattern specification | The process that results in the patterns of cell differentiation that will arise in an embryo. |
| establishment of cell polarity | The specification and formation of anisotropic intracellular organization or cell growth patterns. |
| lateral root development | The process whose specific outcome is the progression of the lateral root over time, from its formation to the mature structure. A lateral root is one formed from pericycle cells located on the xylem radius of the root, as opposed to the initiation of the main root from the embryo proper. |
| protein peptidyl-prolyl isomerization | The modification of a protein by cis-trans isomerization of a proline residue. |
| response to cytokinin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokinin stimulus. |
| root development | The process whose specific outcome is the progression of the root over time, from its formation to the mature structure. The root is the water- and mineral-absorbing part of a plant which is usually underground, does not bear leaves, tends to grow downwards and is typically derived from the radicle of the embryo. |
| unidimensional cell growth | The process in which a cell irreversibly increases in size in one |
| very long-chain fatty acid biosynthetic process | The chemical reactions and pathways resulting in the formation of a fatty acid which has a chain length greater than C22. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A6QQ71 | FKBP6 | Inactive peptidyl-prolyl cis-trans isomerase FKBP6 | Bos taurus (Bovine) | PR |
| Q9W1I9 | shu | Inactive peptidyl-prolyl cis-trans isomerase shutdown | Drosophila melanogaster (Fruit fly) | PR |
| Q14318 | FKBP8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | Homo sapiens (Human) | EV |
| Q02790 | FKBP4 | Peptidyl-prolyl cis-trans isomerase FKBP4 | Homo sapiens (Human) | PR |
| O35465 | Fkbp8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | Mus musculus (Mouse) | SS |
| Q3B7U9 | Fkbp8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAVGDQTEQN | YLPKKKKSET | EDDKRRKKIV | PGSLLKAVVR | PGGGDSSPVD | GDQVIYHCTV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RTLDGVVVES | TRSESGGRGV | PIRDVLGNSK | MILGLLEGIP | TMHKGEIAMF | KMKPEMHYAE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IDCPVSAPEN | FPKDDELHFE | IELLDFSKAK | IASDDLGVIK | KILNEGEGWE | SPREPYEVKA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RISAKSGDGH | VIFSHTEEPY | FFTFGKSEVP | KGLEIGIGTM | ARKEKAVIYV | RKQYLTESPL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LHIDQDLEEV | HFEVELVHFI | QVRDMLGDGR | LIKRRIRDGR | GEFPMDCPLQ | DSRLSVHYKG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MLLNEEKTVF | YDSKIDNNDQ | PLEFSSGEGL | VPEGFEMCTR | LMLPGEIALV | TCPPDYAYDK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FPRPPGVSEG | AHVQWEIELL | GFETPRDWTG | LNFQSIMDEA | DKIRSTGNRL | FKEGKFELAK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AKYEKVLREF | NHVNPQDEDE | GKIFGDTRNM | LHLNVAACLL | KMGEWRKSIE | TCNKVLEAKP |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GHVKGLYRRG | MAYIAGGEYD | DARNDFNMMI | KVDKSSEADA | TAALLKLKQK | EQEAESKARK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| QFKGLFDKRP | GEITEVGSEI | REESKTIEEV | DETKDNDDDE | TLEEEGATTV | STERKRKWSE |
| 610 | 620 | 630 | |||
| KAWPFLKNVM | LQIGIQLGVV | LIGILIFQFV | SAKFT |