Q6ZVM7
Gene name |
TOM1L2 |
Protein name |
TOM1-like protein 2 |
Names |
Target of Myb-like protein 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:146691 |
EC number |
|
Protein Class |
|
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
9-156 (VHS domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

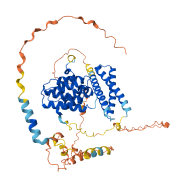
1 structures for Q6ZVM7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6ZVM7-F1 | Predicted | AlphaFoldDB |
523 variants for Q6ZVM7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1322613096 | 5 | L>M | No | gnomAD | |
| rs1388214807 | 7 | N>S | No | gnomAD | |
| rs994462976 | 8 | P>L | No | gnomAD | |
| rs994462976 | 8 | P>R | No | gnomAD | |
| rs2145137730 | 9 | F>S | No | Ensembl | |
| rs2042142733 | 13 | V>L | No | TOPMed | |
| rs1369344607 | 15 | Q>H | No | gnomAD | |
| rs779175296 | 17 | L>F | No |
ExAC TOPMed gnomAD |
|
|
COSM1135887 COSM472375 |
18 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1179016582 | 20 | A>T | No | gnomAD | |
| rs761266752 | 22 | D>G | No |
ExAC gnomAD |
|
| rs1568234298 | 22 | D>N | No | Ensembl | |
| rs1207870258 | 24 | S>F | No | gnomAD | |
| rs145420007 | 24 | S>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1331636916 | 26 | Q>K | No | TOPMed | |
| rs2039151839 | 28 | E>G | No | Ensembl | |
| rs2039151688 | 29 | D>N | No | TOPMed | |
|
COSM1588638 COSM976152 |
31 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772748379 | 31 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1377776005 | 31 | T>S | No |
TOPMed gnomAD |
|
| rs1809263007 | 32 | L>V | No | TOPMed | |
| rs528653796 | 33 | N>I | No |
TOPMed gnomAD |
|
| rs528653796 | 33 | N>T | No |
TOPMed gnomAD |
|
| rs771961539 | 34 | M>T | No |
ExAC gnomAD |
|
| rs759595245 | 35 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs2039150055 | 36 | I>T | No | gnomAD | |
| rs2039149760 | 37 | C>G | No | Ensembl | |
|
rs774380990 COSM3362009 COSM3362008 |
39 | I>T | kidney [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs2039149609 | 39 | I>V | No | TOPMed | |
| rs867591849 | 41 | N>S | No | gnomAD | |
|
COSM5645161 COSM5645160 rs771164302 |
43 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs386352375 CA232177 RCV000122484 |
48 | K>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs1389567152 | 51 | I>V | No | TOPMed | |
| rs139022042 | 52 | R>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1720743 rs867644339 COSM1720744 |
53 | A>V | NS [Cosmic] | No |
cosmic curated Ensembl |
| rs1443147537 | 54 | L>P | No |
TOPMed gnomAD |
|
|
COSM4917167 COSM4917166 |
55 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1277991354 | 56 | K>T | No | gnomAD | |
| rs547421443 | 57 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs867464276 | 57 | R>W | No |
TOPMed gnomAD |
|
| rs1339111033 | 58 | L>F | No |
TOPMed gnomAD |
|
| rs769042756 | 59 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs748480864 | 60 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs759002706 | 62 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs1348950356 COSM5149209 |
62 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM389901 rs746357732 |
64 | Y>C | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs777391955 | 65 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs2038693295 | 66 | E>G | No | Ensembl | |
| rs2038692855 | 68 | M>I | No | TOPMed | |
| rs2144293430 | 68 | M>V | No | Ensembl | |
| TCGA novel | 70 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1388952005 | 72 | T>I | No | gnomAD | |
| rs375848127 | 73 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2144223464 | 79 | K>* | No | Ensembl | |
| rs2144223408 | 82 | G>V | No | Ensembl | |
| rs775641202 | 83 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs371535609 | 83 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs772516795 | 84 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs759751295 | 84 | R>H | No |
ExAC gnomAD |
|
| rs771396440 | 87 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs771396440 | 87 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs749804679 | 89 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1002369021 | 91 | N>S | No | TOPMed | |
| rs905363686 | 92 | R>Q | No |
TOPMed gnomAD |
|
| rs1245254871 | 93 | D>Y | No |
TOPMed gnomAD |
|
| rs2038413623 | 94 | F>V | No | Ensembl | |
| rs746685451 | 95 | I>M | No |
ExAC gnomAD |
|
| rs768263789 | 95 | I>V | No |
ExAC gnomAD |
|
| rs1427544991 | 96 | D>G | No | gnomAD | |
| rs779535298 | 96 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs2038412564 | 97 | S>R | No | TOPMed | |
| rs2038412306 | 99 | L>V | No | Ensembl | |
| rs1191923763 | 100 | V>A | No |
TOPMed gnomAD |
|
| rs1267197992 | 102 | I>T | No | gnomAD | |
| rs758219410 | 102 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs750130496 | 103 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs778757730 | 103 | I>M | No |
ExAC gnomAD |
|
| rs1598282423 | 103 | I>T | No | Ensembl | |
| rs750130496 | 103 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs375361037 | 104 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs2038409724 | 105 | P>L | No | Ensembl | |
| rs1201854247 | 107 | N>H | No | gnomAD | |
| rs1247326711 | 107 | N>S | No | gnomAD | |
| rs930406175 | 108 | N>D | No |
TOPMed gnomAD |
|
| rs753745234 | 108 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs764180348 | 109 | P>H | No |
ExAC gnomAD |
|
| rs1265849696 | 110 | P>L | No |
TOPMed gnomAD |
|
| rs760704865 | 110 | P>S | No |
ExAC gnomAD |
|
| rs767575657 | 112 | I>V | No |
ExAC gnomAD |
|
| rs1464114114 | 113 | V>A | No |
TOPMed gnomAD |
|
| rs1568189086 | 114 | Q>E | No | Ensembl | |
| rs759787784 | 115 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs2038407532 | 115 | D>Y | No | TOPMed | |
| rs774642358 | 116 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs771387359 | 116 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs771387359 | 116 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs2144220874 | 122 | Q>* | No | Ensembl | |
| TCGA novel | 122 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755576657 | 127 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs755576657 | 127 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs779521366 | 127 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs763374689 | 128 | F>L | No |
ExAC TOPMed gnomAD |
|
|
rs773862235 COSM976148 COSM1588639 |
129 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1003640600 COSM1588640 COSM976147 |
129 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| rs1464712946 | 130 | S>R | No | TOPMed | |
| rs774932395 | 132 | P>A | No |
ExAC gnomAD |
|
| rs2037906298 | 133 | D>A | No |
TOPMed gnomAD |
|
| rs745557175 | 134 | L>F | No |
ExAC gnomAD |
|
| rs2037905815 | 135 | T>A | No | gnomAD | |
| rs903590011 | 135 | T>S | No | Ensembl | |
| rs1568164106 | 136 | G>S | No |
TOPMed gnomAD |
|
|
COSM1658928 COSM1658929 rs200120270 |
137 | V>I | salivary_gland Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs777529542 | 138 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1322878085 | 139 | H>R | No | gnomAD | |
| rs1188970007 | 139 | H>Y | No | TOPMed | |
| rs151155378 | 140 | I>L | No |
ESP ExAC TOPMed |
|
| rs376386507 | 140 | I>M | No | Ensembl | |
| rs151155378 | 140 | I>V | No |
ESP ExAC TOPMed |
|
| TCGA novel | 140 | I>Y | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2037903976 | 141 | Y>H | No | gnomAD | |
| rs1436067803 | 142 | E>K | No | gnomAD | |
| rs931615840 | 145 | K>E | No | Ensembl | |
| rs748084321 | 148 | G>R | No |
ExAC gnomAD |
|
| rs143220830 | 149 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| COSM285864 | 150 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751763566 | 150 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs368309703 | 151 | F>I | No |
ESP ExAC gnomAD |
|
| rs2037902157 | 152 | P>S | No | TOPMed | |
| rs1012287828 | 153 | M>V | No |
1000Genomes TOPMed |
|
| rs374413505 | 154 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs765694471 | 155 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1211442725 | 156 | L>S | No |
TOPMed gnomAD |
|
| rs139869743 | 157 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1488653951 | 157 | D>N | No | gnomAD | |
| rs983267767 | 158 | A>T | No | gnomAD | |
| rs766958630 | 159 | L>M | No |
ExAC gnomAD |
|
|
COSM4064420 COSM4064419 |
160 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1326103673 | 160 | S>F | No | gnomAD | |
| rs2037899434 | 162 | I>M | No | Ensembl | |
| rs1316428930 | 162 | I>R | No | gnomAD | |
| rs774005970 | 163 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1489894555 | 164 | T>S | No | gnomAD | |
| rs770625527 | 165 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs2144091084 | 165 | P>L | No | 1000Genomes | |
| rs770625527 | 165 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs773028858 | 167 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs773028858 | 167 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1027041802 | 167 | R>W | No | gnomAD | |
| rs1278376881 | 170 | P>A | No | gnomAD | |
| rs772799062 | 172 | V>G | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs762640105 |
172 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD NCI-TCGA |
| rs762640105 | 172 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1164955701 | 173 | D>A | No |
TOPMed gnomAD |
|
| rs2037801047 | 173 | D>E | No | Ensembl | |
| rs142581994 | 173 | D>N | No |
ESP TOPMed gnomAD |
|
| rs769503926 | 174 | P>L | No |
ExAC gnomAD |
|
| rs148462085 | 174 | P>S | No | ESP | |
| rs770869564 | 175 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs776451196 | 176 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2037799579 | 177 | T>I | No | Ensembl | |
| rs1305956046 | 178 | M>I | No |
TOPMed gnomAD |
|
| rs2037798971 | 178 | M>T | No | gnomAD | |
| rs2144061715 | 178 | M>V | No | Ensembl | |
| rs746942840 | 179 | P>S | No |
ExAC gnomAD |
|
|
COSM1381253 COSM1381254 rs1270944554 |
180 | R>G | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs2037798182 | 180 | R>T | No | Ensembl | |
| rs144591677 | 181 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs2037797367 | 184 | Q>K | No | TOPMed | |
| rs779157435 | 185 | Q>H | No |
ExAC gnomAD |
|
| rs1352648192 | 186 | R>G | No | gnomAD | |
| rs1352648192 | 186 | R>W | No | gnomAD | |
| rs200715176 | 188 | S>N | No | Ensembl | |
| rs149335483 | 189 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1038317699 | 190 | G>S | No | Ensembl | |
| rs2037796088 | 192 | Y>C | No |
TOPMed gnomAD |
|
| rs2037795937 | 193 | S>Y | No | TOPMed | |
| rs199563013 | 194 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1442094533 | 194 | S>T | No | gnomAD | |
| rs756682926 | 195 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs138147144 | 195 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs762405956 | 196 | P>L | No |
ExAC gnomAD |
|
| rs370279147 | 197 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2037794548 | 197 | P>S | No | Ensembl | |
| rs764890724 | 198 | A>S | No |
ExAC gnomAD |
|
| rs2037794070 | 198 | A>V | No | gnomAD | |
| rs1382920456 | 199 | P>R | No |
TOPMed gnomAD |
|
| rs776559462 | 200 | Y>C | No |
ExAC gnomAD |
|
| rs1239092795 | 200 | Y>H | No | gnomAD | |
| rs1356221251 | 201 | S>A | No |
TOPMed gnomAD |
|
| rs1461782638 | 201 | S>F | No | gnomAD | |
| rs775520074 | 202 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs775520074 | 202 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2037792113 | 202 | A>V | No | Ensembl | |
| rs745885112 | 203 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs745885112 | 203 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs745885112 | 203 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs866923102 | 205 | A>V | No | Ensembl | |
| rs139158330 | 206 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs139158330 | 206 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
|
rs929538302 COSM1664628 COSM1664629 |
206 | P>S | kidney [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs756419453 | 207 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs756419453 | 207 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs2037789131 | 209 | S>C | No | TOPMed | |
| rs748681398 | 210 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs142515051 | 213 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs146710625 | 213 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs146710625 | 213 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs77916160 | 214 | I>N | No | gnomAD | |
| rs77916160 | 214 | I>T | No | gnomAD | |
| rs756977667 | 214 | I>V | No |
ExAC gnomAD |
|
| rs1163706261 | 215 | T>P | No | gnomAD | |
| rs753411232 | 217 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs763971752 | 218 | S>* | No |
ExAC gnomAD |
|
| rs760507746 | 219 | E>Q | No |
ExAC gnomAD |
|
| rs941795814 | 219 | E>V | No | gnomAD | |
| rs1205255326 | 220 | Q>R | No | TOPMed | |
| rs1294492433 | 221 | I>M | No | gnomAD | |
| rs1028789047 | 221 | I>T | No | Ensembl | |
| rs753560204 | 222 | A>S | No |
ExAC gnomAD |
|
| rs753560204 | 222 | A>T | No |
ExAC gnomAD |
|
| rs763773589 | 222 | A>V | No |
ExAC gnomAD |
|
| rs767388904 | 225 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs752605260 | 225 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2037615138 | 226 | S>N | No | TOPMed | |
| rs1431708325 | 228 | L>V | No | gnomAD | |
| TCGA novel | 229 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369984263 | 230 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs773493334 | 231 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs143816562 | 232 | R>* | No |
ESP ExAC gnomAD |
|
| rs1480281935 | 232 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs762021018 | 234 | N>S | No |
ExAC gnomAD |
|
| rs777024931 | 236 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs768989668 | 237 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs1202059213 | 238 | M>I | No | gnomAD | |
| rs1326829489 | 238 | M>V | No | gnomAD | |
| rs2037612872 | 239 | S>P | No | gnomAD | |
| rs552793390 | 239 | S>Y | No |
1000Genomes ExAC gnomAD |
|
| rs1411562529 | 240 | E>K | No | gnomAD | |
|
COSM3932497 COSM3932496 |
240 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1329504819 | 241 | M>I | No | TOPMed | |
| rs1263321281 | 241 | M>L | No | gnomAD | |
| rs1321064158 | 243 | T>A | No |
TOPMed gnomAD |
|
| rs770481247 | 243 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs765936175 | 244 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1391882632 | 244 | E>K | No |
TOPMed gnomAD |
|
| rs1391882632 | 244 | E>Q | No |
TOPMed gnomAD |
|
| rs752374687 | 248 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs752374687 | 248 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs755869161 | 248 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs752374687 | 248 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1175431247 | 249 | Q>* | No |
TOPMed gnomAD |
|
| rs2037610345 | 249 | Q>H | No | TOPMed | |
| rs1468445668 | 250 | E>K | No | gnomAD | |
| rs926275423 | 251 | D>Y | No | Ensembl | |
| rs1177338439 | 252 | S>* | No | gnomAD | |
| rs2037609870 | 252 | S>P | No | TOPMed | |
| rs781156044 | 253 | S>P | No |
ExAC gnomAD |
|
| rs1162063872 | 256 | E>D | No | gnomAD | |
| rs1192505143 | 257 | L>S | No | TOPMed | |
| rs766168764 | 258 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2037039836 | 260 | E>G | No | TOPMed | |
| rs1297928954 | 261 | L>I | No | gnomAD | |
| rs1568112760 | 262 | N>K | No |
TOPMed gnomAD |
|
| rs764167124 | 262 | N>S | No |
ExAC gnomAD |
|
| rs760939410 | 263 | R>K | No |
ExAC gnomAD |
|
| rs1598221611 | 264 | T>P | No | Ensembl | |
| rs1365705483 | 265 | C>G | No | gnomAD | |
| rs767900512 | 266 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs775756759 | 266 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs759982924 | 267 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2046762602 | 269 | Q>* | No | TOPMed | |
| rs769430448 | 270 | Q>E | No |
ExAC gnomAD |
|
| rs769430448 | 270 | Q>K | No |
ExAC gnomAD |
|
| rs2037037030 | 270 | Q>P | No | TOPMed | |
|
COSM3818954 COSM3818953 |
271 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2143841974 | 271 | R>S | No | Ensembl | |
| rs1357291759 | 272 | I>M | No | gnomAD | |
| rs747599352 | 273 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1206594445 | 274 | E>V | No |
TOPMed gnomAD |
|
| rs1053760343 | 275 | L>F | No |
TOPMed gnomAD |
|
| rs2037035322 | 276 | I>L | No | TOPMed | |
| rs768256975 | 276 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1598221339 | 277 | S>P | No | Ensembl | |
| rs2037034797 | 277 | S>Y | No | TOPMed | |
| rs578191280 | 278 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs578191280 | 278 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1303005080 COSM1230064 COSM1230063 |
278 | R>H | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs779766547 | 279 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1568111972 | 281 | N>H | No | gnomAD | |
| rs537305430 | 281 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1370940561 | 282 | E>* | No | gnomAD | |
| rs1201339578 | 282 | E>G | No | gnomAD | |
| rs1480610218 | 283 | E>* | No |
TOPMed gnomAD |
|
| rs2037032322 | 284 | V>D | No |
TOPMed gnomAD |
|
| rs199832973 | 285 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs764201570 | 286 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs371612900 | 287 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs752915063 | 290 | H>R | No |
ExAC gnomAD |
|
| rs1225740489 | 291 | V>M | No | gnomAD | |
| rs2037029927 | 293 | D>H | No | Ensembl | |
| rs2037029927 | 293 | D>N | No | Ensembl | |
| rs2037029544 | 294 | D>N | No | Ensembl | |
| TCGA novel | 294 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs536434387 | 296 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4064418 COSM4064417 rs1034038096 |
298 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs2037028553 | 299 | F>L | No |
TOPMed gnomAD |
|
| rs746666657 | 300 | L>F | No |
ExAC gnomAD |
|
| rs745605701 | 301 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs745605701 | 301 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs745605701 | 301 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs145438922 | 302 | Y>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1026905220 | 302 | Y>* | No | TOPMed | |
| TCGA novel | 302 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778667027 | 302 | Y>H | No |
ExAC gnomAD |
|
| rs1364271756 | 303 | E>D | No | gnomAD | |
| rs547369838 | 303 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547369838 | 303 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2036879817 | 305 | F>I | No | gnomAD | |
| TCGA novel | 305 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759206629 | 306 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs372470867 | 307 | R>* | No |
ESP TOPMed gnomAD |
|
| rs770717113 | 307 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs770717113 | 307 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2036878554 | 308 | Y>S | No | Ensembl | |
| rs762750960 | 309 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1315595947 | 309 | R>K | No |
TOPMed gnomAD |
|
| rs1315595947 | 309 | R>M | No |
TOPMed gnomAD |
|
| rs930707964 | 309 | R>S | No |
TOPMed gnomAD |
|
| rs769833356 | 312 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs748001425 | 312 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2036876293 | 313 | S>C | No | TOPMed | |
| TCGA novel | 313 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1287368983 | 313 | S>P | No |
TOPMed gnomAD |
|
| rs768809501 | 314 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs768809501 | 314 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1362445170 | 315 | Q>H | No |
TOPMed gnomAD |
|
| rs747269852 | 317 | A>G | No |
ExAC gnomAD |
|
| TCGA novel | 317 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs899598049 | 318 | S>G | No | TOPMed | |
| TCGA novel | 318 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1303614072 | 319 | N>D | No | gnomAD | |
| rs2143800995 | 320 | G>A | No | Ensembl | |
| rs747032744 | 323 | N>D | No |
ExAC gnomAD |
|
| rs775698263 | 323 | N>I | No |
ExAC gnomAD |
|
| rs746047482 | 323 | N>K | No |
ExAC gnomAD |
|
| rs2036852315 | 326 | T>S | No | gnomAD | |
| rs757520450 | 327 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs757520450 | 327 | E>Q | No |
ExAC gnomAD |
|
| rs576894860 | 328 | D>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2036851561 | 329 | N>S | No | TOPMed | |
| TCGA novel | 334 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1192074612 | 336 | G>V | No | TOPMed | |
| rs1471172897 | 337 | S>Y | No | TOPMed | |
| rs369187930 | 339 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs751132404 | 340 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1310393787 | 342 | S>I | No | gnomAD | |
| rs758116852 | 344 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs749992298 | 346 | G>R | No |
ExAC gnomAD |
|
| rs146446650 | 347 | N>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs761451871 | 348 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs375083600 | 349 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2036848014 | 350 | P>L | No | gnomAD | |
| rs775753326 | 351 | P>S | No |
ExAC gnomAD |
|
|
COSM3514814 rs978993333 COSM3514813 |
353 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs2036847066 | 354 | L>P | No | Ensembl | |
| rs1161907104 | 355 | S>F | No | gnomAD | |
| rs1357929540 | 355 | S>P | No | gnomAD | |
| rs772024120 | 358 | L>P | No |
ExAC gnomAD |
|
| rs1176443207 | 360 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1176443207 | 360 | G>V | No | gnomAD | |
| rs1598213131 | 361 | L>I | No | TOPMed | |
| rs1598213131 | 361 | L>V | No | TOPMed | |
| rs763953806 | 364 | G>E | No |
ExAC gnomAD |
|
| rs753756874 | 364 | G>R | No |
ExAC gnomAD |
|
| rs752635967 | 366 | E>D | No |
ExAC gnomAD |
|
| rs760746771 | 366 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs144061121 | 367 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs144061121 | 367 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2036633405 | 368 | V>A | No | gnomAD | |
| rs774472699 | 368 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs774472699 | 368 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs774472699 | 368 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs763131283 | 369 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs763131283 | 369 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs2036632886 | 370 | G>D | No | TOPMed | |
| rs773614320 | 371 | T>N | No |
ExAC TOPMed gnomAD |
|
|
COSM1588643 COSM976144 |
374 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2036632407 | 376 | Q>K | No | TOPMed | |
| rs2036632242 | 377 | Q>E | No | TOPMed | |
| rs139141212 | 378 | C>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs897638691 | 381 | R>C | No |
TOPMed gnomAD |
|
|
COSM976143 rs375549455 COSM1153033 |
381 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs375549455 | 381 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs757014791 | 382 | D>G | No |
ExAC gnomAD |
|
| rs1050660895 | 383 | G>A | No |
TOPMed gnomAD |
|
| rs1232150695 | 383 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
COSM976142 COSM1588644 |
385 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1477334598 | 386 | M>T | No | gnomAD | |
| rs1267623734 | 386 | M>V | No |
TOPMed gnomAD |
|
| rs1261020499 | 388 | A>D | No | gnomAD | |
| rs563051881 | 390 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs563051881 | 390 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2036628576 | 394 | S>Y | No | gnomAD | |
| rs2036628405 | 395 | L>F | No |
TOPMed gnomAD |
|
| rs1486206873 | 397 | E>G | No | TOPMed | |
|
COSM4064413 COSM4064414 rs763182884 |
399 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs770151031 COSM3514811 COSM3514812 |
399 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs770151031 | 399 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1489425999 | 400 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs574247504 | 400 | K>M | No |
1000Genomes ExAC gnomAD |
|
| rs574247504 | 400 | K>R | No |
1000Genomes ExAC gnomAD |
|
|
COSM4064412 COSM4064411 |
401 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769111096 | 401 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs867950133 | 404 | Y>C | No |
TOPMed gnomAD |
|
| rs867950133 | 404 | Y>F | No |
TOPMed gnomAD |
|
| rs1338194353 | 404 | Y>H | No |
TOPMed gnomAD |
|
| rs1323137249 | 406 | D>G | No |
TOPMed gnomAD |
|
| rs1261447179 | 407 | P>S | No |
TOPMed gnomAD |
|
| rs1261447179 | 407 | P>T | No |
TOPMed gnomAD |
|
| rs761112529 | 408 | Q>H | No |
ExAC gnomAD |
|
| rs748862321 | 411 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1029007401 | 412 | G>R | No | TOPMed | |
| rs772840957 | 416 | A>V | No |
ExAC gnomAD |
|
| rs2036558433 | 418 | D>E | No | TOPMed | |
| rs2036558262 | 419 | N>D | No | gnomAD | |
| rs974715100 | 419 | N>S | No |
TOPMed gnomAD |
|
| rs1385530930 | 420 | R>* | No |
TOPMed gnomAD |
|
|
COSM316072 COSM1588645 rs769606765 |
420 | R>Q | lung Variant assessed as Somatic; MODERATE impact. endometrium [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs747813834 | 422 | Q>R | No |
ExAC gnomAD |
|
| rs1185640622 | 425 | E>V | No |
TOPMed gnomAD |
|
| rs958547512 | 426 | G>A | No |
TOPMed gnomAD |
|
| rs2035939554 | 427 | I>L | No | Ensembl | |
| rs756282247 | 427 | I>N | No |
ExAC gnomAD |
|
|
COSM4064407 COSM4064408 rs150784633 |
429 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1302469 COSM1302468 rs1209314216 |
430 | A>V | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs2035938135 | 431 | Q>* | No | TOPMed | |
| rs775992105 | 432 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs775992105 | 432 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs972472710 | 435 | M>L | No |
TOPMed gnomAD |
|
| rs972472710 | 435 | M>V | No |
TOPMed gnomAD |
|
| rs775314255 | 437 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1568045038 | 438 | I>T | No | Ensembl | |
| rs1268895163 | 438 | I>V | No |
TOPMed gnomAD |
|
| rs1381767405 | 440 | V>M | No | gnomAD | |
| rs771808887 | 441 | W>C | No |
ExAC gnomAD |
|
| rs1568044943 | 441 | W>S | No | Ensembl | |
| rs372626043 | 442 | L>F | No |
ESP ExAC gnomAD |
|
| TCGA novel | 442 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770932579 | 445 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs770932579 | 445 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1214475621 | 448 | G>C | No | gnomAD | |
| rs1224315397 | 449 | D>G | No | TOPMed | |
| rs1489596178 | 449 | D>Y | No | gnomAD | |
| rs747446015 | 452 | E>K | No |
ExAC gnomAD |
|
| rs2035802411 | 453 | E>D | No | gnomAD | |
| rs1320275314 | 453 | E>K | No |
TOPMed gnomAD |
|
| rs1280705025 | 454 | G>R | No |
TOPMed gnomAD |
|
| rs1280705025 | 454 | G>S | No |
TOPMed gnomAD |
|
| rs1219751199 | 454 | G>V | No |
TOPMed gnomAD |
|
| rs926879365 | 456 | T>K | No |
TOPMed gnomAD |
|
| rs2035801112 | 457 | S>T | No | TOPMed | |
| rs200566614 | 458 | E>D | No |
TOPMed gnomAD |
|
| rs1174232191 | 461 | D>H | No |
TOPMed gnomAD |
|
| rs1467689959 | 462 | K>R | No | gnomAD | |
| rs1467689959 | 462 | K>T | No | gnomAD | |
| rs1378246708 | 465 | E>D | No | gnomAD | |
| TCGA novel | 467 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1198204191 | 467 | R>K | No | gnomAD | |
| rs1354242684 | 468 | A>G | No |
TOPMed gnomAD |
|
| rs766989418 | 469 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs200243623 | 471 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs908488984 | 474 | V>A | No | TOPMed | |
| rs908488984 | 474 | V>D | No | TOPMed | |
| rs751129069 | 475 | P>A | No |
ExAC gnomAD |
|
| rs766199692 | 475 | P>R | No |
ExAC gnomAD |
|
| rs1598162302 | 476 | D>A | No | Ensembl | |
| rs761812249 | 476 | D>E | No |
ExAC gnomAD |
|
| rs761903481 | 476 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs2035727447 | 477 | L>F | No |
TOPMed gnomAD |
|
| rs768807403 | 478 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs768807403 | 478 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1230358121 | 478 | P>S | No | gnomAD | |
| rs370628988 | 479 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
| rs1442039712 | 480 | P>S | No | TOPMed | |
| rs2143428660 | 481 | P>A | No | Ensembl | |
| rs746278350 | 481 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs780707839 | 482 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1568033566 | 482 | M>L | No | Ensembl | |
| rs749744673 | 482 | M>R | No |
ExAC gnomAD |
|
| rs749744673 | 482 | M>T | No |
ExAC gnomAD |
|
| rs1382475008 | 483 | E>K | No | gnomAD | |
| rs2035723883 | 484 | A>D | No | Ensembl | |
| rs1439525637 | 484 | A>S | No |
TOPMed gnomAD |
|
| rs1231557027 | 485 | P>S | No | gnomAD | |
| rs1443507502 | 486 | A>P | No |
TOPMed gnomAD |
|
| rs202079010 | 487 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs750224529 | 488 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2143427256 | 488 | A>V | No | Ensembl | |
| rs765104318 | 492 | S>F | No |
ExAC gnomAD |
|
| rs2035721993 | 493 | G>A | No |
TOPMed gnomAD |
|
| rs761867100 | 493 | G>S | No |
ExAC gnomAD |
|
| rs2035721993 | 493 | G>V | No |
TOPMed gnomAD |
|
| rs371456416 | 494 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs371456416 | 494 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1679614 COSM1679613 rs539750130 |
494 | R>W | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs775631393 | 495 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs2035721277 | 495 | K>Q | No | gnomAD | |
| TCGA novel | 497 | P>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1022295863 | 498 | E>G | No | Ensembl | |
| rs1332115184 | 499 | R>P | No |
TOPMed gnomAD |
|
|
COSM1381250 rs1332115184 COSM1381251 |
499 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs772252818 | 499 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs774716816 | 501 | E>A | No |
ExAC gnomAD |
|
| rs1419348668 | 501 | E>D | No |
TOPMed gnomAD |
|
| rs2035719286 | 502 | D>V | No | TOPMed | |
| rs1332145908 | 503 | A>V | No |
TOPMed gnomAD |
|
| rs2035718540 | 504 | L>F | No | TOPMed | |
| rs778194590 | 505 | F>V | No |
ExAC gnomAD |
|
| rs746566696 | 506 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1263313471 | 506 | A>V | No | gnomAD | |
| rs886209868 | 507 | L>P | No | gnomAD | |
| rs886209868 | 507 | L>R | No | gnomAD | |
| rs200990292 | 507 | L>V | No |
ExAC TOPMed gnomAD |
No associated diseases with Q6ZVM7
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| clathrin binding | Binding to a clathrin heavy or light chain, the main components of the coat of coated vesicles and coated pits, and which also occurs in synaptic vesicles. |
| phosphatidylinositol binding | Binding to an inositol-containing glycerophospholipid, i.e. phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| ubiquitin binding | Binding to ubiquitin, a protein that when covalently bound to other cellular proteins marks them for proteolytic degradation. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| negative regulation of mitotic nuclear division | Any process that stops, prevents or reduces the rate or extent of mitosis. Mitosis is the division of the eukaryotic cell nucleus to produce two daughter nuclei that, usually, contain the identical chromosome complement to their mother. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9NZ52 | GGA3 | ADP-ribosylation factor-binding protein GGA3 | Homo sapiens (Human) | EV |
| O75674 | TOM1L1 | TOM1-like protein 1 | Homo sapiens (Human) | SS |
| Q8BMI3 | Gga3 | ADP-ribosylation factor-binding protein GGA3 | Mus musculus (Mouse) | SS |
| O88746 | Tom1 | Target of Myb1 membrane trafficking protein | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEFLLGNPFS | TPVGQCLEKA | TDGSLQSEDW | TLNMEICDII | NETEEGPKDA | IRALKKRLNG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NRNYREVMLA | LTVLETCVKN | CGHRFHILVA | NRDFIDSVLV | KIISPKNNPP | TIVQDKVLAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IQAWADAFRS | SPDLTGVVHI | YEELKRKGVE | FPMADLDALS | PIHTPQRSVP | EVDPAATMPR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SQSQQRTSAG | SYSSPPPAPY | SAPQAPALSV | TGPITANSEQ | IARLRSELDV | VRGNTKVMSE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| MLTEMVPGQE | DSSDLELLQE | LNRTCRAMQQ | RIVELISRVS | NEEVTEELLH | VNDDLNNVFL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RYERFERYRS | GRSVQNASNG | VLNEVTEDNL | IDLGPGSPAV | VSPMVGNTAP | PSSLSSQLAG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LDLGTESVSG | TLSSLQQCNP | RDGFDMFAQT | RGNSLAEQRK | TVTYEDPQAV | GGLASALDNR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KQSSEGIPVA | QPSVMDDIEV | WLRTDLKGDD | LEEGVTSEEF | DKFLEERAKA | AEMVPDLPSP |
| 490 | 500 | ||||
| PMEAPAPASN | PSGRKKPERS | EDALFAL |