Q6QNF3
Gene name |
PDGFRB (PDGFR, PDGFR1) |
Protein name |
Platelet-derived growth factor receptor beta |
Names |
PDGF-R-beta , PDGFR-beta , EC 2.7.10.1 , Beta platelet-derived growth factor receptor , Beta-type platelet-derived growth factor receptor , CD140 antigen-like family member B , Platelet-derived growth factor receptor 1 , PDGFR-1 , CD antigen CD140b |
Species |
Canis lupus familiaris (Dog) (Canis familiaris) |
KEGG Pathway |
cfa:442985 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
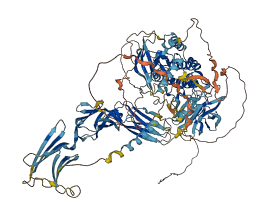
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
600-962 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Target domain |
600-962 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Hubbard SR (2004) "Juxtamembrane autoinhibition in receptor tyrosine kinases", Nature reviews. Molecular cell biology, 5, 464-71
- Chiara F et al. (2004) "Autoinhibition of the platelet-derived growth factor beta-receptor tyrosine kinase by its C-terminal tail", The Journal of biological chemistry, 279, 19732-8
Autoinhibited structure

Activated structure

1 structures for Q6QNF3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6QNF3-F1 | Predicted | AlphaFoldDB |
No variants for Q6QNF3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q6QNF3 | |||||
No associated diseases with Q6QNF3
16 regional properties for Q6QNF3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 600 - 962 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 601 - 955 | IPR001245 |
| conserved_site | Tyrosine-protein kinase, receptor class III, conserved site | 659 - 672 | IPR001824 |
| domain | Immunoglobulin subtype 2 | 45 - 107 | IPR003598-1 |
| domain | Immunoglobulin subtype 2 | 226 - 298 | IPR003598-2 |
| domain | Immunoglobulin subtype 2 | 331 - 403 | IPR003598-3 |
| domain | Immunoglobulin subtype | 39 - 121 | IPR003599-1 |
| domain | Immunoglobulin subtype | 220 - 311 | IPR003599-2 |
| domain | Immunoglobulin subtype | 325 - 414 | IPR003599-3 |
| domain | Immunoglobulin-like domain | 20 - 101 | IPR007110-1 |
| domain | Immunoglobulin-like domain | 214 - 307 | IPR007110-2 |
| active_site | Tyrosine-protein kinase, active site | 822 - 834 | IPR008266 |
| domain | Immunoglobulin I-set | 334 - 413 | IPR013098 |
| domain | Immunoglobulin-like beta-sandwich domain | 40 - 109 | IPR013151 |
| binding_site | Protein kinase, ATP binding site | 606 - 634 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 600 - 958 | IPR020635 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| lysosomal lumen | The volume enclosed within the lysosomal membrane. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| platelet-derived growth factor beta-receptor activity | Combining with platelet-derived growth factor isoform PDGF-BB or PDGF-AB to initiate a change in cell activity. |
| platelet-derived growth factor binding | Binding to platelet-derived growth factor. |
17 GO annotations of biological process
| Name | Definition |
|---|---|
| cell chemotaxis | The directed movement of a motile cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| cell migration involved in coronary angiogenesis | The orderly movement of a cell from one site to another that will contribute to the formation of new blood vessels in the heart from pre-existing blood vessels. |
| cell migration involved in vasculogenesis | The orderly movement of a cell from one site to another that will contribute to the differentiation of an endothelial cell that will form de novo blood vessels and tubes. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| phosphatidylinositol metabolic process | The chemical reactions and pathways involving phosphatidylinositol, any glycophospholipid in which a sn-glycerol 3-phosphate residue is esterified to the 1-hydroxyl group of 1D-myo-inositol. |
| platelet-derived growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a platelet-derived growth factor receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| positive regulation of calcium-mediated signaling | Any process that activates or increases the frequency, rate or extent of calcium-mediated signaling. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of ERK1 and ERK2 cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| positive regulation of phospholipase C activity | Any process that increases the rate of phospholipase C activity. |
| positive regulation of phosphoprotein phosphatase activity | Any process that activates or increases the activity of a phosphoprotein phosphatase. |
| positive regulation of smooth muscle cell migration | Any process that activates, maintains or increases the frequency, rate or extent of smooth muscle cell migration. |
| positive regulation of smooth muscle cell proliferation | Any process that activates or increases the rate or extent of smooth muscle cell proliferation. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| retina vasculature development in camera-type eye | The process whose specific outcome is the progression of the vasculature of the retina over time, from its formation to the mature structure. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q75ZY9 | MET | Hepatocyte growth factor receptor | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| O97799 | KIT | Mast/stem cell growth factor receptor Kit | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P09619 | PDGFRB | Platelet-derived growth factor receptor beta | Homo sapiens (Human) | EV |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| Q05030 | Pdgfrb | Platelet-derived growth factor receptor beta | Rattus norvegicus (Rat) | SS |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LDQ3 | CRK34 | Cysteine-rich receptor-like protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQVPGTMPAP | VLKGQALWLP | LLLMLSPQAS | GGLVITPPGP | ELVLNISSTF | VLTCSGPAPV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VWERLSQEPL | QKMARTQDGT | FSSTLTLTNV | TGLDTGEYFC | TYKGSHGLEP | DGRKRLYIFV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PDPTMGFLPV | DPEELFIFLT | EITEITIPCR | VTDPRLVVTL | HEKKVDIPLP | IPYDHQRGFS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GTFEDKTYVC | KTTIGDKEVD | SEAYYVYSLQ | VSSINVSVNA | VQTVVRQGEN | ITIMCIVTGN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EVVNFEWTYP | RMESGRLVEP | VTDFLFNVPS | HIRSILHIPS | AELGDSGTYI | CNVSESVNDH |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RDEKSINVTV | VESGYVRLLG | ELDAVQFAEL | HRSRALQVVF | EAYPPPTVVW | FKDNRTLGDS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SAGEIALSTR | NVSETRYVSE | LTLVRVKVAE | AGYYTMRAFH | EDAEAQLSFQ | LQVNVPVRVL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ELSESHPASG | EQTVRCRGRG | MPQPHLTWST | CSDLKRCPRE | LPPTPLGNSS | EEESQLETNV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| TYWPEDQEFE | VVSTLRLRRV | DQPLSVRCTL | HNLLGHDMQE | VTVVPHSLPF | KVVVISAILA |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LVVLTIISLI | ILIMLWQKKP | RYEIRWKVIE | SVSSDGHEYI | YVDPMQLPYD | STWELPRDQL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| VLGRTLGSGA | FGQVVEATAH | GLSHSQATMK | VAVKMLKSTA | RSSEKQALMS | ELKIMSHLGP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HLNVVNLLGA | CTKGGPIYII | TEYCRYGDLV | DYLHRNKHTF | LQLCSDKRRP | PSAELYSNAL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| PAGLPLPSHV | SLPGESDGGY | MDMSKDESVD | YVPMLDMKGG | VKYADIESSS | YMAPYDNYVP |
| 790 | 800 | 810 | 820 | 830 | 840 |
| TAPERTCRAT | LINESPVLSY | TDLVGFSYQV | ANGMEFLASK | NCVHRDLAAR | NVLICEGKLV |
| 850 | 860 | 870 | 880 | 890 | 900 |
| KICDFGLARD | IMRDSNYISK | GSTFLPLKWM | APESIFNSLY | TTLSDVWSFG | ILLWEIFTLG |
| 910 | 920 | 930 | 940 | 950 | 960 |
| GTPYPELPMN | EQFYNAIKRG | YRMAQPAHAS | DEIYEIMQKC | WEEKFEIRPP | FSQLVLLLER |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| LLGEGYKKKY | QQVDEEFLRS | DHPAVLRSQA | RLPGFPGLRS | PLDTSSVLYT | AVQPNEGDND |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| YIIPLPDPKP | EVADGPLESS | PSLASSTLNE | VNTSSTISCD | SPLEPQEEPE | PEPEPQPEPQ |
| 1090 | 1100 | ||||
| VVPEPPLDSS | CPGPRAEAED | SFL |