Q6P9R2
Gene name |
Oxsr1 |
Protein name |
Serine/threonine-protein kinase OSR1 |
Names |
Oxidative stress-responsive 1 protein |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:108737 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
163-191 (Activation loop from InterPro)
Target domain |
17-291 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
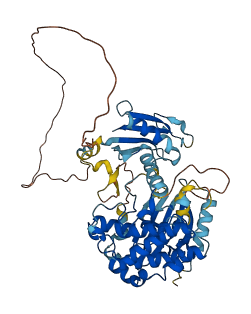
Autoinhibited structure

Activated structure

1 structures for Q6P9R2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6P9R2-F1 | Predicted | AlphaFoldDB |
31 variants for Q6P9R2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389082561 | 56 | S>R | No | EVA | |
| rs3400172383 | 77 | S>Y | No | EVA | |
| rs3400196323 | 78 | Y>* | No | EVA | |
| rs3400534594 | 79 | Y>D | No | EVA | |
| rs3389087284 | 90 | L>I | No | EVA | |
| rs3389074752 | 109 | A>S | No | EVA | |
| rs3389077619 | 138 | H>P | No | EVA | |
| rs3389081183 | 152 | I>V | No | EVA | |
| rs3389090871 | 159 | S>T | No | EVA | |
| rs3389090864 | 204 | Y>C | No | EVA | |
| rs3389087313 | 213 | F>L | No | EVA | |
| rs3400196301 | 261 | G>A | No | EVA | |
| rs3389077562 | 269 | S>L | No | EVA | |
| rs3389091719 | 271 | C>S | No | EVA | |
| rs3399645639 | 301 | Q>* | No | EVA | |
| rs3389060898 | 311 | I>N | No | EVA | |
| rs3389049569 | 321 | V>I | No | EVA | |
| rs3389074708 | 346 | E>K | No | EVA | |
| rs3389068878 | 349 | E>K | No | EVA | |
| rs3389077537 | 360 | P>H | No | EVA | |
| rs236012427 | 368 | S>N | No | EVA | |
| rs585287900 | 373 | A>S | No | EVA | |
| rs3389086010 | 375 | A>T | No | EVA | |
| rs223116630 | 390 | A>V | No | EVA | |
| rs227124595 | 420 | A>V | No | EVA | |
| rs3389049624 | 456 | P>H | No | EVA | |
| rs3389026724 | 458 | R>G | No | EVA | |
| rs3389077586 | 474 | V>A | No | EVA | |
| rs3389057458 | 486 | L>M | No | EVA | |
| rs3389082627 | 488 | K>M | No | EVA | |
| rs3389090842 | 498 | S>Y | No | EVA |
No associated diseases with Q6P9R2
7 regional properties for Q6P9R2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | WD40 repeat | 216 - 258 | IPR001680-1 |
| repeat | WD40 repeat | 261 - 300 | IPR001680-2 |
| repeat | WD40 repeat | 355 - 402 | IPR001680-3 |
| repeat | WD40 repeat | 459 - 507 | IPR001680-4 |
| repeat | WD40 repeat | 564 - 605 | IPR001680-5 |
| repeat | WD40 repeat | 844 - 886 | IPR001680-6 |
| conserved_site | WD40 repeat, conserved site | 380 - 394 | IPR019775 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| identical protein binding | Binding to an identical protein or proteins. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
15 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular hypotonic response | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of detection of, or exposure to, a hypotonic environment, i.e. an environment with a lower concentration of solutes than the organism or cell. |
| cellular response to chemokine | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemokine stimulus. |
| chemokine (C-C motif) ligand 21 signaling pathway | The series of molecular signals initiated by the binding of the C-C chemokine CCL21 to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| chemokine (C-X-C motif) ligand 12 signaling pathway | The series of molecular signals initiated by the binding of the chemokine CXCL12 to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of potassium ion transmembrane transport | Any process that stops, prevents or reduces the frequency, rate or extent of potassium ion transmembrane transport. |
| negative regulation of potassium ion transmembrane transporter activity | Any process that stops, prevents or reduces the frequency, rate or extent of potassium ion transmembrane transporter activity. |
| osmosensory signaling pathway | The series of molecular signals initiated in response to osmotic change. |
| peptidyl-threonine phosphorylation | The phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
| positive regulation of T cell chemotaxis | Any process that increases the rate, frequency or extent of T cell chemotaxis. T cell chemotaxis is the directed movement of a T cell in response to an external stimulus. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| response to oxidative stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| response to xenobiotic stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a xenobiotic, a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
20 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9C0K7 | STRADB | STE20-related kinase adapter protein beta | Homo sapiens (Human) | PR |
| Q7RTN6 | STRADA | STE20-related kinase adapter protein alpha | Homo sapiens (Human) | PR |
| Q9UEW8 | STK39 | STE20/SPS1-related proline-alanine-rich protein kinase | Homo sapiens (Human) | PR |
| O95747 | OXSR1 | Serine/threonine-protein kinase OSR1 | Homo sapiens (Human) | PR |
| Q8K4T3 | Stradb | STE20-related kinase adapter protein beta | Mus musculus (Mouse) | PR |
| Q3UUJ4 | Strada | STE20-related kinase adapter protein alpha | Mus musculus (Mouse) | PR |
| Q9Z1W9 | Stk39 | STE20/SPS1-related proline-alanine-rich protein kinase | Mus musculus (Mouse) | PR |
| O54988 | Slk | STE20-like serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| O55098 | Stk10 | Serine/threonine-protein kinase 10 | Mus musculus (Mouse) | PR |
| P70218 | Map4k1 | Mitogen-activated protein kinase kinase kinase kinase 1 | Mus musculus (Mouse) | PR |
| Q61161 | Map4k2 | Mitogen-activated protein kinase kinase kinase kinase 2 | Mus musculus (Mouse) | SS |
| Q99JP0 | Map4k3 | Mitogen-activated protein kinase kinase kinase kinase 3 | Mus musculus (Mouse) | SS |
| Q99JT2 | Stk26 | Serine/threonine-protein kinase 26 | Mus musculus (Mouse) | PR |
| Q99KH8 | Stk24 | Serine/threonine-protein kinase 24 | Mus musculus (Mouse) | PR |
| Q9Z2W1 | Stk25 | Serine/threonine-protein kinase 25 | Mus musculus (Mouse) | PR |
| Q8BPM2 | Map4k5 | Mitogen-activated protein kinase kinase kinase kinase 5 | Mus musculus (Mouse) | PR |
| Q863I2 | OXSR1 | Serine/threonine-protein kinase OSR1 | Sus scrofa (Pig) | PR |
| Q7TNZ6 | Strada | STE20-related kinase adapter protein alpha | Rattus norvegicus (Rat) | PR |
| O88506 | Stk39 | STE20/SPS1-related proline-alanine-rich protein kinase | Rattus norvegicus (Rat) | PR |
| O23304 | BLUS1 | Serine/threonine-protein kinase BLUS1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSEDSSALPW | SINRDDYELQ | EVIGSGATAV | VQAAYCAPKK | ERVAIKRINL | EKCQTSMDEL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LKEIQAMSQC | HHPNIVSYYT | SFVVKDELWL | VMKLLSGGSV | LDIIKHIVAK | GEHKSGVLDE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PTIATILREV | LEGLEYLHKN | GQIHRDVKAG | NILLGEDGSV | QIADFGVSAF | LATGGDITRN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KVRKTFVGTP | CWMAPEVMEQ | VRGYDFKADI | WSFGITAIEL | ATGAAPYHKY | PPMKVLMLTL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QNDPPSLETG | VQDKEMLKKY | GKSFRKMISL | CLQKDPEKRP | TAAELLRHKF | FQKAKNKEFL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| QEKILQRAPT | ISERSKKVRR | VPGSSGRLHK | TEDGGWEWSD | DEFDEESEEG | RAAISQLRSP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RVKDSLSSSE | LFAAAEPMGT | LLQVPEQISA | HLPQPAGQMP | TQPAQVSLLP | PAEPAKPAQA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QSSGERSQET | KIPISLVLRL | RNSKKELNDI | RFEFTPGRDT | AEGVSQELIS | AGLVDGRDLV |
| 490 | 500 | 510 | 520 | ||
| IVAANLQKIV | EEPQSNRSVT | FKLASGVEGS | DIPDDGKLIG | FAQLSIS |