Q6P2M8
Gene name |
PNCK |
Protein name |
Calcium/calmodulin-dependent protein kinase type 1B |
Names |
EC 2.7.11.17 , CaM kinase I beta , CaM kinase IB , CaM-KI beta , CaMKI-beta , Pregnancy up-regulated non-ubiquitously-expressed CaM kinase |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:139728 |
EC number |
2.7.11.17: Protein-serine/threonine kinases |
Protein Class |
|
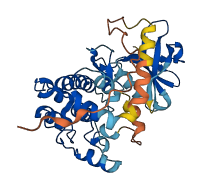
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
15-270 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
156-177 (Activation loop from InterPro)
Target domain |
11-287 (Catalytic domain of the Serine/Threonine kinase, Calcium/calmodulin-dependent protein kinase Type I beta) |
Relief mechanism |
|
Assay |
|
156-177 (Activation loop from InterPro)
Target domain |
11-287 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Goldberg J et al. (1996) "Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I", Cell, 84, 875-87
- Yuan T et al. (2004) "Spectroscopic characterization of the calmodulin-binding and autoinhibitory domains of calcium/calmodulin-dependent protein kinase I", Archives of biochemistry and biophysics, 421, 192-206
- Clapperton JA et al. (2002) "Structure of the complex of calmodulin with the target sequence of calmodulin-dependent protein kinase I: studies of the kinase activation mechanism", Biochemistry, 41, 14669-79
- Zha M et al. (2012) "Crystal structures of human CaMKIα reveal insights into the regulation mechanism of CaMKI", PloS one, 7, e44828
Autoinhibited structure

Activated structure

1 structures for Q6P2M8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6P2M8-F1 | Predicted | AlphaFoldDB |
197 variants for Q6P2M8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA415092211 rs1342350201 |
7 | H>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs782507126 CA10549064 |
11 | I>S | No |
ClinGen ExAC gnomAD |
|
|
rs371353486 CA10549063 |
12 | S>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA415092135 rs1349409547 |
18 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1202463497 CA415092133 |
18 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA415092137 rs1349409547 |
18 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1557040655 CA415092131 |
19 | E>K | No |
ClinGen gnomAD |
|
|
rs782381672 CA10549062 |
20 | R>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs146604878 CA337242434 |
21 | L>V | No |
ClinGen ESP |
|
|
COSM1599042 CA10549061 COSM1599043 rs782551985 |
23 | S>L | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA |
|
rs1557040418 CA415092088 |
24 | G>R | No |
ClinGen gnomAD |
|
|
CA10549004 rs782249570 COSM3780384 COSM3780385 |
28 | E>K | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
COSM1145644 CA415092010 COSM1651053 rs1557040387 |
35 | R>Q | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1409549653 CA415092012 |
35 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA415091981 rs144452893 |
38 | A>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10548999 rs144452893 |
38 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs144452893 CA10548998 |
38 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs782773576 CA10548997 |
39 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA415091905 rs1213167326 |
45 | C>F | No |
ClinGen TOPMed |
|
|
rs1282358274 CA415091850 |
50 | A>V | No |
ClinGen TOPMed |
|
|
rs1557040353 CA415091832 |
52 | R>Q | No |
ClinGen gnomAD |
|
|
CA10548994 rs368559736 |
52 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs1557040340 CA415091827 |
53 | G>D | No |
ClinGen gnomAD |
|
|
CA415091788 rs1557040326 |
56 | A>D | No |
ClinGen gnomAD |
|
|
rs1569538728 CA415091768 |
59 | E>Q | No |
ClinGen Ensembl |
|
|
CA10548991 rs782729326 |
61 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781976704 CA10548990 |
63 | A>T | Variant assessed as Somatic; 8.401e-05 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA10548989 rs139296709 |
66 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA10548988 rs782395857 |
66 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1557040225 CA415091517 |
70 | H>Q | No |
ClinGen gnomAD |
|
|
COSM3843972 COSM3843973 CA415091507 rs1388242444 |
71 | P>L | Variant assessed as Somatic; impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs782660842 CA10548954 |
73 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA10548952 rs781791539 |
74 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA415091457 rs1557040202 |
75 | A>S | No |
ClinGen gnomAD |
|
|
CA415091459 rs1557040202 |
75 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1557040198 CA415091438 |
76 | L>R | No |
ClinGen gnomAD |
|
|
rs782100269 CA10548950 RCV000626119 |
77 | E>K | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA337241743 rs923490329 |
78 | D>G | No |
ClinGen Ensembl |
|
|
CA10548949 rs781858294 |
79 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415091388 rs1557040179 |
82 | S>T | No |
ClinGen gnomAD |
|
|
rs1557040175 CA415091363 |
85 | H>N | No |
ClinGen gnomAD |
|
|
CA337241731 rs964811956 |
86 | L>F | No |
ClinGen Ensembl |
|
| TCGA novel | 93 | V>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10548924 rs375555808 |
94 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA10548922 rs782155273 |
96 | G>D | No |
ClinGen ExAC |
|
|
CA415090292 rs1465664873 |
97 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA10548919 rs199567613 |
101 | R>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs200243657 CA10548918 |
101 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed |
|
|
rs782376200 CA10548916 |
103 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1557040082 CA415090215 |
103 | M>L | No |
ClinGen gnomAD |
|
|
CA10548917 rs781952916 |
103 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs782253314 CA10548915 |
105 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA337241559 rs934422979 |
105 | R>L | No |
ClinGen gnomAD |
|
|
CA415090176 rs1286264111 |
106 | G>S | No |
ClinGen TOPMed |
|
|
rs782667539 CA10548914 |
109 | T>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 110 | E>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs147245837 CA10548913 |
112 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1223977507 CA415090067 |
114 | S>G | No |
ClinGen TOPMed |
|
|
CA10548912 rs782175890 |
114 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA337241532 rs781794626 |
115 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
CA10548911 rs782581121 |
115 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs782459101 CA10548910 |
116 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1557040048 CA415090020 |
118 | G>V | No |
ClinGen gnomAD |
|
|
rs1050798291 CA337241523 |
120 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
rs781844576 CA10548909 |
123 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA10548908 rs782759815 |
123 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs782109329 CA10548907 |
127 | L>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA415089935 rs1179167704 |
130 | L>P | No |
ClinGen TOPMed |
|
|
COSM1557144 CA10548905 COSM1557145 rs782815240 |
131 | G>W | lung Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA |
|
CA415089907 rs1557040008 |
133 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA415089877 rs143688303 |
135 | R>P | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA337241471 rs143688303 |
135 | R>Q | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA415089881 COSM1315323 rs1557039999 COSM1315324 |
135 | R>W | Variant assessed as Somatic; 0.0 impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1375952238 CA415089866 |
136 | D>V | No |
ClinGen TOPMed |
|
|
CA415089857 rs369888785 |
137 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10548903 rs369888785 |
137 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA415089783 rs1468161459 |
139 | P>L | No |
ClinGen TOPMed |
|
|
rs1255168600 CA415089748 |
142 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA415089729 rs1557039891 |
144 | Y>H | No |
ClinGen gnomAD |
|
|
rs782768077 CA10548884 |
145 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA10548883 rs782141197 |
146 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA415089698 rs1557039883 |
147 | P>S | No |
ClinGen gnomAD |
|
|
rs782061174 CA10548880 |
150 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA415089634 rs1192586225 |
151 | S>W | No |
ClinGen TOPMed |
|
|
CA415089620 rs1169370262 |
152 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1460789436 CA415089599 |
154 | M>K | No |
ClinGen TOPMed |
|
|
rs1603201137 CA415089602 |
154 | M>V | No |
ClinGen Ensembl |
|
|
rs201553565 CA337241132 |
157 | D>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA10548877 rs201553565 |
157 | D>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1165160719 CA415089530 |
159 | G>A | No |
ClinGen TOPMed |
|
|
rs1557039851 CA415089520 |
160 | L>F | No |
ClinGen gnomAD |
|
|
rs782409266 CA10548875 |
168 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA10548873 COSM1145642 COSM1651057 rs782561753 |
172 | A>T | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA415089304 rs1557039842 |
178 | Y>F | No |
ClinGen gnomAD |
|
|
CA10548832 rs782201125 |
183 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs781963016 CA10548831 |
185 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA10548830 rs782365846 |
186 | Q>* | No |
ClinGen ExAC |
|
|
CA415089076 rs868928121 |
188 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA10548828 rs377279617 |
189 | Y>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10548826 rs782189775 |
190 | G>W | No |
ClinGen ExAC gnomAD |
|
|
CA415089057 rs1569538603 |
191 | K>T | No |
ClinGen Ensembl |
|
|
rs1375299700 CA415089050 |
192 | A>P | No |
ClinGen TOPMed |
|
|
CA10548824 rs782154493 |
193 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415089022 rs1557039723 |
196 | W>* | No |
ClinGen gnomAD |
|
|
CA337240756 rs782013647 |
200 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs782413389 CA10548813 |
206 | L>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 208 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782214506 CA10548809 |
210 | P>R | No |
ClinGen ExAC gnomAD |
|
| rs781891121 | 212 | F>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781891121 | 212 | F>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1176043604 CA415088898 |
214 | D>N | No |
ClinGen TOPMed |
|
|
CA10548803 rs782670579 |
215 | E>K | No |
ClinGen ExAC |
|
|
rs1461755892 CA415088875 |
217 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1299131323 CA415088852 |
220 | L>I | No |
ClinGen TOPMed |
|
|
CA10548801 rs781807763 |
222 | S>N | No |
ClinGen ExAC |
|
| TCGA novel | 223 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1398825868 CA415088828 |
223 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA415088808 rs1325552815 |
226 | R>T | No |
ClinGen TOPMed |
|
|
CA10548800 rs782600758 |
228 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs781849317 CA10548798 |
230 | E>V | No |
ClinGen ExAC gnomAD |
|
|
rs1557039622 CA415088774 |
231 | F>S | No |
ClinGen gnomAD |
|
| TCGA novel | 232 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10548797 rs782787717 |
233 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
CA415088706 rs1236647063 |
239 | I>V | No |
ClinGen TOPMed |
|
|
rs782421620 CA10548788 |
247 | I>N | No |
ClinGen ExAC gnomAD |
|
|
CA415088530 rs1171497590 |
248 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
COSM1466575 CA337240476 COSM1466576 rs915970109 |
248 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA415088462 rs1557039562 |
252 | E>K | No |
ClinGen gnomAD |
|
|
CA10548787 rs782172793 |
253 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA415088424 rs1557039557 |
253 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA415088396 rs1164445052 |
254 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA415088411 rs1460270465 |
254 | D>H | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 255 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782591480 CA10548786 |
255 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA415088336 rs1557039545 |
258 | R>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA415088304 rs1395517968 |
259 | F>L | No |
ClinGen TOPMed |
|
|
CA10548785 rs782336468 |
259 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA415088266 rs1557039537 |
261 | C>Y | No |
ClinGen gnomAD |
|
|
CA337240445 VAR_040598 rs56060609 |
262 | Q>H | No |
ClinGen UniProt 1000Genomes ExAC dbSNP gnomAD |
|
|
rs200742175 CA10548784 |
262 | Q>R | No |
ClinGen 1000Genomes ExAC TOPMed |
|
|
CA415088221 rs1359704059 |
263 | Q>R | No |
ClinGen TOPMed |
|
|
rs1381566915 CA415088192 |
264 | A>V | No |
ClinGen TOPMed |
|
|
CA10548780 rs782668687 |
266 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA415088174 rs781882136 |
266 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10548778 rs781812862 |
269 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA10548755 rs782234685 |
274 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs782342775 CA337240308 |
274 | T>I | No |
ClinGen Ensembl |
|
|
rs1557039466 CA415087964 |
275 | A>D | No |
ClinGen gnomAD |
|
|
rs1557039462 CA415087937 |
277 | D>H | No |
ClinGen gnomAD |
|
|
COSM1599050 rs1557039462 COSM1599049 CA415087939 |
277 | D>N | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 278 | R>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557039459 CA415087827 |
281 | L>S | No |
ClinGen gnomAD |
|
|
CA415087780 rs1210538448 |
283 | S>F | No |
ClinGen TOPMed |
|
|
CA10548751 rs141426834 |
285 | S>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10548750 rs782450206 |
288 | I>M | No |
ClinGen ExAC |
|
|
rs146644423 CA10548748 |
289 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10548749 rs181550205 |
289 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10548747 rs145782812 |
291 | N>D | No |
ClinGen ESP ExAC |
|
|
rs1557039440 CA415087658 |
291 | N>S | No |
ClinGen gnomAD |
|
|
COSM3235684 rs782789099 CA10548745 COSM3235685 |
294 | R>Q | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs371383897 CA10548746 |
294 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1557039431 CA415087592 |
295 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA415087579 rs1362308788 |
296 | H>Q | No |
ClinGen TOPMed |
|
|
CA337240271 rs1046700989 |
297 | W>* | No |
ClinGen Ensembl |
|
|
CA10548722 rs782746502 |
299 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782111633 CA10548721 |
299 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA415087410 rs1557039361 |
302 | N>S | No |
ClinGen gnomAD |
|
| TCGA novel | 304 | T>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415087350 rs1557039358 |
305 | S>L | No |
ClinGen gnomAD |
|
|
rs782008969 CA10548720 |
308 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10548719 rs782198402 |
308 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
COSM3406104 COSM3406103 CA10548716 rs782337569 |
311 | R>Q | Variant assessed as Somatic; 0.0 impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA10548717 rs781926333 |
311 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs148971524 CA10548715 |
314 | G>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA415087157 rs1308507964 |
317 | P>L | No |
ClinGen TOPMed |
|
|
CA10548714 rs782723734 |
319 | G>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA415087122 rs1251419103 |
320 | E>D | No |
ClinGen TOPMed |
|
|
CA415087134 rs1557039309 |
320 | E>K | No |
ClinGen gnomAD |
|
| TCGA novel | 321 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1468074812 CA415087105 |
322 | A>T | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 322 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557039285 CA415087057 |
325 | Q>R | No |
ClinGen Ensembl |
|
| TCGA novel | 326 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557039279 CA415087040 |
326 | G>V | No |
ClinGen gnomAD |
|
|
rs782688994 CA10548711 |
327 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781912274 CA10548710 |
328 | A>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA10548709 rs782441624 |
329 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10548708 rs782492056 |
329 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10548707 rs782492056 |
329 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781855747 CA10548706 |
330 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA337240022 rs1005287908 |
335 | L>F | No |
ClinGen Ensembl |
|
|
CA10548703 rs7890468 |
336 | R>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs782711198 CA10548702 |
336 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs147664846 CA337240007 |
340 | P>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs147664846 CA10548701 |
340 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1557039245 | 342 | K>missing | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557039240 CA415086822 |
343 | W>* | No |
ClinGen gnomAD |
No associated diseases with Q6P2M8
11 regional properties for Q6P2M8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Dbl homology (DH) domain | 535 - 726 | IPR000219 |
| domain | FERM domain | 44 - 324 | IPR000299 |
| domain | Pleckstrin homology domain | 755 - 854 | IPR001849-1 |
| domain | Pleckstrin homology domain | 929 - 1028 | IPR001849-2 |
| domain | FERM adjacent | 332 - 378 | IPR014847 |
| domain | FERM, N-terminal | 48 - 110 | IPR018979 |
| domain | FERM, C-terminal PH-like domain | 238 - 328 | IPR018980 |
| conserved_site | FERM conserved site | 98 - 127 | IPR019747 |
| domain | FERM central domain | 129 - 234 | IPR019748 |
| domain | Band 4.1 domain | 40 - 234 | IPR019749 |
| domain | FARP1/FARP2/FRMD7, FERM domain C-lobe | 221 - 341 | IPR041788 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.17 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions |
| protein serine kinase activity | Catalysis of the reactions |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
106 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4IFM7 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Bos taurus (Bovine) | SS |
| Q0V7M1 | PSKH1 | Serine/threonine-protein kinase H1 | Bos taurus (Bovine) | SS |
| Q0VD22 | STK33 | Serine/threonine-protein kinase 33 | Bos taurus (Bovine) | PR |
| O15075 | DCLK1 | Serine/threonine-protein kinase DCLK1 | Homo sapiens (Human) | EV |
| Q9UIK4 | DAPK2 | Death-associated protein kinase 2 | Homo sapiens (Human) | EV |
| O94768 | STK17B | Serine/threonine-protein kinase 17B | Homo sapiens (Human) | PR |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| Q8N568 | DCLK2 | Serine/threonine-protein kinase DCLK2 | Homo sapiens (Human) | PR |
| Q9UEE5 | STK17A | Serine/threonine-protein kinase 17A | Homo sapiens (Human) | SS |
| O75962 | TRIO | Triple functional domain protein | Homo sapiens (Human) | EV |
| Q8WWF8 | CAPSL | Calcyphosin-like protein | Homo sapiens (Human) | PR |
| P53355 | DAPK1 | Death-associated protein kinase 1 | Homo sapiens (Human) | EV |
| Q9BYT3 | STK33 | Serine/threonine-protein kinase 33 | Homo sapiens (Human) | PR |
| O43293 | DAPK3 | Death-associated protein kinase 3 | Homo sapiens (Human) | PR |
| Q16816 | PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | Homo sapiens (Human) | PR |
| P15735 | PHKG2 | Phosphorylase b kinase gamma catalytic chain, liver/testis isoform | Homo sapiens (Human) | PR |
| Q13554 | CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | Homo sapiens (Human) | EV |
| Q9UQM7 | CAMK2A | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Homo sapiens (Human) | EV |
| Q13555 | CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Homo sapiens (Human) | EV |
| Q13557 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Homo sapiens (Human) | EV |
| Q9H1R3 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Homo sapiens (Human) | EV |
| Q32MK0 | MYLK3 | Myosin light chain kinase 3 | Homo sapiens (Human) | SS |
| Q86YV6 | MYLK4 | Myosin light chain kinase family member 4 | Homo sapiens (Human) | SS |
| P11801 | PSKH1 | Serine/threonine-protein kinase H1 | Homo sapiens (Human) | SS |
| Q96NX5 | CAMK1G | Calcium/calmodulin-dependent protein kinase type 1G | Homo sapiens (Human) | SS |
| Q8IU85 | CAMK1D | Calcium/calmodulin-dependent protein kinase type 1D | Homo sapiens (Human) | SS |
| Q8NCB2 | CAMKV | CaM kinase-like vesicle-associated protein | Homo sapiens (Human) | SS |
| Q14012 | CAMK1 | Calcium/calmodulin-dependent protein kinase type 1 | Homo sapiens (Human) | EV |
| Q91VB2 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Mus musculus (Mouse) | SS |
| Q8BG48 | Stk17b | Serine/threonine-protein kinase 17B | Mus musculus (Mouse) | PR |
| Q8VCR8 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Mus musculus (Mouse) | SS |
| Q6PGN3 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Mus musculus (Mouse) | PR |
| Q91YS8 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Mus musculus (Mouse) | SS |
| Q3UIZ8 | Mylk3 | Myosin light chain kinase 3 | Mus musculus (Mouse) | SS |
| Q8VDF3 | Dapk2 | Death-associated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q6P8Y1 | Capsl | Calcyphosin-like protein | Mus musculus (Mouse) | PR |
| Q8BW96 | Camk1d | Calcium/calmodulin-dependent protein kinase type 1D | Mus musculus (Mouse) | SS |
| Q3UHL1 | Camkv | CaM kinase-like vesicle-associated protein | Mus musculus (Mouse) | SS |
| P08414 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Mus musculus (Mouse) | SS |
| Q9JLM8 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Mus musculus (Mouse) | SS |
| Q91YA2 | Pskh1 | Serine/threonine-protein kinase H1 | Mus musculus (Mouse) | SS |
| Q80YE7 | Dapk1 | Death-associated protein kinase 1 | Mus musculus (Mouse) | SS |
| Q62407 | Speg | Striated muscle-specific serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| Q0KL02 | Trio | Triple functional domain protein | Mus musculus (Mouse) | SS |
| Q924X7 | Stk33 | Serine/threonine-protein kinase 33 | Mus musculus (Mouse) | PR |
| O54784 | Dapk3 | Death-associated protein kinase 3 | Mus musculus (Mouse) | PR |
| Q9QYK9 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Mus musculus (Mouse) | PR |
| Q7TNJ7 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Rattus norvegicus (Rat) | SS |
| P20689 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Rattus norvegicus (Rat) | SS |
| F1M0Z1 | Trio | Triple functional domain protein | Rattus norvegicus (Rat) | SS |
| Q91XS8 | Stk17b | Serine/threonine-protein kinase 17B | Rattus norvegicus (Rat) | PR |
| O08875 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Rattus norvegicus (Rat) | PR |
| E9PT87 | Mylk3 | Myosin light chain kinase 3 | Rattus norvegicus (Rat) | SS |
| Q63092 | Camkv | CaM kinase-like vesicle-associated protein | Rattus norvegicus (Rat) | SS |
| Q63638 | Speg | Striated muscle-specific serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| O88764 | Dapk3 | Death-associated protein kinase 3 | Rattus norvegicus (Rat) | PR |
| P13234 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Rattus norvegicus (Rat) | SS |
| Q63450 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Rattus norvegicus (Rat) | EV |
| P97924 | Kalrn | Kalirin | Rattus norvegicus (Rat) | SS |
| Q5MPA9 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Rattus norvegicus (Rat) | PR |
| O70150 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Rattus norvegicus (Rat) | PR |
| Q6AVM3 | CCAMK | Calcium and calcium/calmodulin-dependent serine/threonine-protein kinase | Oryza sativa subsp japonica (Rice) | PR |
| P53682 | CPK23 | Calcium-dependent protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| P53684 | CPK7 | Calcium-dependent protein kinase 7 | Oryza sativa subsp japonica (Rice) | PR |
| Q5VQQ5 | CPK2 | Calcium-dependent protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6I5I8 | CPK16 | Calcium-dependent protein kinase 16 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6K968 | CPK6 | Calcium-dependent protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75GE8 | CPK8 | Calcium-dependent protein kinase 8 | Oryza sativa subsp. japonica (Rice) | SS |
| Q84SL0 | CPK20 | Calcium-dependent protein kinase 20 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LPZ7 | CPK3 | Calcium-dependent protein kinase 3 | Oryza sativa subsp. japonica (Rice) | SS |
| O44997 | dapk-1 | Death-associated protein kinase dapk-1 | Caenorhabditis elegans | SS |
| Q95QC4 | zyg-8 | Serine/threonine-protein kinase zyg-8 | Caenorhabditis elegans | PR |
| Q9TXJ0 | cmk-1 | Calcium/calmodulin-dependent protein kinase type 1 | Caenorhabditis elegans | PR |
| P28583 | Calcium-dependent protein kinase SK5 | Glycine max (Soybean) (Glycine hispida) | PR | |
| Q9M9V8 | CPK10 | Calcium-dependent protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FX86 | CRK8 | CDPK-related kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSF8 | CPK30 | Calcium-dependent protein kinase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIQ7 | CPK24 | Calcium-dependent protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93759 | GK-1 | Calcium-dependent protein kinase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8S8S2 | LPEAT2 | Lysophospholipid acyltransferase LPEAT2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LJL9 | CRK2 | CDPK-related kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SG12 | CRK6 | CDPK-related kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCS2 | CRK5 | CDPK-related kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4I7 | CPK13 | Calcium-dependent protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NLQ6 | CPK32 | Calcium-dependent protein kinase 32 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38873 | CPK7 | Calcium-dependent protein kinase 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42438 | CPK8 | Calcium-dependent protein kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1LUA6 | trio | Triple functional domain protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A8C984 | mylk3 | Myosin light chain kinase 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q7SY49 | camkv | CaM kinase-like vesicle-associated protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLLLKKHTED | ISSVYEIRER | LGSGAFSEVV | LAQERGSAHL | VALKCIPKKA | LRGKEALVEN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EIAVLRRISH | PNIVALEDVH | ESPSHLYLAM | ELVTGGELFD | RIMERGSYTE | KDASHLVGQV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LGAVSYLHSL | GIVHRDLKPE | NLLYATPFED | SKIMVSDFGL | SKIQAGNMLG | TACGTPGYVA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PELLEQKPYG | KAVDVWALGV | ISYILLCGYP | PFYDESDPEL | FSQILRASYE | FDSPFWDDIS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESAKDFIRHL | LERDPQKRFT | CQQALRHLWI | SGDTAFDRDI | LGSVSEQIRK | NFARTHWKRA |
| 310 | 320 | 330 | 340 | ||
| FNATSFLRHI | RKLGQIPEGE | GASEQGMARH | SHSGLRAGQP | PKW |