Q6P1E8
Gene name |
Efcab6 (Djbp, Kiaa1672) |
Protein name |
EF-hand calcium-binding domain-containing protein 6 |
Names |
DJ-1-binding protein, DJBP |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:77627 |
EC number |
|
Protein Class |
|
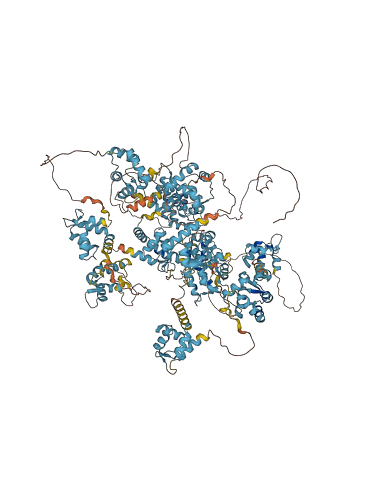
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for Q6P1E8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 8I7O | EM | 450 A | Q1/Q2 | 1-1516 | PDB |
| 8I7R | EM | 650 A | Q1/Q2/Q3 | 1-1516 | PDB |
| 8IYJ | EM | 350 A | i2/i3/i4 | 1-1516 | PDB |
| 8TO0 | EM | 770 A | Ez | 1-1516 | PDB |
| AF-Q6P1E8-F1 | Predicted | AlphaFoldDB |
96 variants for Q6P1E8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs263171788 | 8 | L>P | No | EVA | |
| rs214116925 | 31 | S>A | No | EVA | |
| rs250786682 | 41 | S>P | No | EVA | |
| rs3389379632 | 60 | R>S | No | EVA | |
| rs232589423 | 75 | A>T | No | EVA | |
| rs248683068 | 91 | A>S | No | EVA | |
| rs3389330687 | 113 | N>I | No | EVA | |
| rs31736732 | 115 | M>T | No | EVA | |
| rs3389393781 | 136 | Q>H | No | EVA | |
| rs265027590 | 165 | I>T | No | EVA | |
| rs259212768 | 184 | R>G | No | EVA | |
| rs31737467 | 186 | V>L | No | EVA | |
| rs229668983 | 197 | R>Q | No | EVA | |
| rs3389387760 | 204 | K>* | No | EVA | |
| rs3406237360 | 228 | T>P | No | EVA | |
| rs3389375337 | 246 | N>I | No | EVA | |
| rs249839079 | 247 | I>L | No | EVA | |
| rs3389349034 | 248 | D>N | No | EVA | |
| rs215450906 | 256 | N>I | No | EVA | |
| rs31734013 | 293 | L>P | No | EVA | |
| rs3389359848 | 323 | Y>* | No | EVA | |
| rs31738570 | 362 | F>L | No | EVA | |
| rs3389330734 | 372 | K>* | No | EVA | |
| rs3389387713 | 383 | L>P | No | EVA | |
| rs3389372946 | 390 | Q>R | No | EVA | |
| rs3389372977 | 402 | K>Q | No | EVA | |
| rs3389359831 | 423 | K>R | No | EVA | |
| rs3389381844 | 433 | K>N | No | EVA | |
| rs3389369114 | 465 | D>N | No | EVA | |
| rs3389359817 | 496 | T>I | No | EVA | |
| rs3389330682 | 543 | R>S | No | EVA | |
| rs3389371864 | 553 | F>I | No | EVA | |
| rs3389371868 | 596 | G>S | No | EVA | |
| rs3389387711 | 597 | I>M | No | EVA | |
| rs231342178 | 601 | T>I | No | EVA | |
| rs265620645 | 603 | P>L | No | EVA | |
| rs247173540 | 607 | P>L | No | EVA | |
| rs222422144 | 617 | I>T | No | EVA | |
| rs253800643 | 624 | D>G | No | EVA | |
| rs260597553 | 629 | P>S | No | EVA | |
| rs242103681 | 632 | E>D | No | EVA | |
| rs3389359880 | 634 | T>N | No | EVA | |
| rs223535677 | 637 | I>L | No | EVA | |
| rs31737196 | 648 | G>S | No | EVA | |
| rs238244436 | 658 | P>S | No | EVA | |
| rs3389381872 | 696 | Y>N | No | EVA | |
| rs214494304 | 697 | A>S | No | EVA | |
| rs3389381902 | 712 | S>N | No | EVA | |
| rs221186929 | 726 | S>R | No | EVA | |
| rs253901214 | 741 | T>M | No | EVA | |
| rs242405118 | 742 | N>S | No | EVA | |
| rs258758502 | 788 | F>L | No | EVA | |
| rs3389342121 | 803 | E>D | No | EVA | |
| rs3389375288 | 850 | A>G | No | EVA | |
| rs3389372969 | 851 | D>G | No | EVA | |
| rs3389387766 | 855 | A>V | No | EVA | |
| rs3389297661 | 874 | L>M | No | EVA | |
| rs215056133 | 932 | H>Q | No | EVA | |
| rs3389297673 | 943 | H>P | No | EVA | |
| rs3389381740 | 948 | E>G | No | EVA | |
| rs3389379674 | 976 | R>K | No | EVA | |
| rs3389372983 | 1048 | S>N | No | EVA | |
| rs31735605 | 1054 | R>H | No | EVA | |
| rs218467944 | 1054 | R>S | No | EVA | |
| rs232036884 | 1071 | V>L | No | EVA | |
| rs216001749 | 1093 | E>K | No | EVA | |
| rs1132934753 | 1096 | S>F | No | EVA | |
| rs1132437054 | 1098 | L>M | No | EVA | |
| rs1135092143 | 1099 | D>E | No | EVA | |
| rs229244810 | 1101 | E>V | No | EVA | |
| rs3389371846 | 1109 | M>V | No | EVA | |
| rs215723472 | 1161 | D>E | No | EVA | |
| rs31730165 | 1161 | D>N | No | EVA | |
| rs3389393722 | 1170 | P>T | No | EVA | |
| rs254126348 | 1173 | M>T | No | EVA | |
| rs31730164 | 1181 | R>K | No | EVA | |
| rs3389378557 | 1196 | H>Y | No | EVA | |
| rs230184911 | 1210 | S>N | No | EVA | |
| rs3389375327 | 1213 | V>F | No | EVA | |
| rs3389297676 | 1250 | K>T | No | EVA | |
| rs233629151 | 1278 | M>I | No | EVA | |
| rs3389378555 | 1282 | G>E | No | EVA | |
| rs3389379649 | 1287 | E>D | No | EVA | |
| rs3389378585 | 1317 | V>E | No | EVA | |
| rs237932408 | 1352 | T>M | No | EVA | |
| rs3389372941 | 1413 | S>Y | No | EVA | |
| rs247449125 | 1419 | R>K | No | EVA | |
| rs3389371830 | 1428 | E>D | No | EVA | |
| rs3389393793 | 1446 | K>M | No | EVA | |
| rs238682504 | 1463 | K>E | No | EVA | |
| rs3389393726 | 1467 | G>* | No | EVA | |
| rs3389387802 | 1484 | N>K | No | EVA | |
| rs3389369111 | 1486 | S>* | No | EVA | |
| rs3389387758 | 1497 | Y>* | No | EVA | |
| rs3389369092 | 1509 | D>E | No | EVA | |
| rs3389372949 | 1515 | L>R | No | EVA |
No associated diseases with Q6P1E8
17 regional properties for Q6P1E8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | EF-hand domain | 96 - 131 | IPR002048-1 |
| domain | EF-hand domain | 197 - 260 | IPR002048-2 |
| domain | EF-hand domain | 325 - 353 | IPR002048-3 |
| domain | EF-hand domain | 444 - 493 | IPR002048-4 |
| domain | EF-hand domain | 532 - 560 | IPR002048-5 |
| domain | EF-hand domain | 659 - 687 | IPR002048-6 |
| domain | EF-hand domain | 763 - 798 | IPR002048-7 |
| domain | EF-hand domain | 905 - 940 | IPR002048-8 |
| domain | EF-hand domain | 1086 - 1121 | IPR002048-9 |
| domain | EF-hand domain | 1197 - 1225 | IPR002048-10 |
| domain | EF-hand domain | 1342 - 1373 | IPR002048-11 |
| domain | EF-hand domain | 1453 - 1514 | IPR002048-12 |
| domain | DJBP, EF-hand domain | 1177 - 1281 | IPR015070 |
| binding_site | EF-Hand 1, calcium-binding site | 109 - 121 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 210 - 222 | IPR018247-2 |
| binding_site | EF-Hand 1, calcium-binding site | 776 - 788 | IPR018247-3 |
| binding_site | EF-Hand 1, calcium-binding site | 1462 - 1474 | IPR018247-4 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q80X60 | Efcab3 | EF-hand calcium-binding domain-containing protein 3 | Mus musculus (Mouse) | PR |
| Q66HC0 | Efcab3 | EF-hand calcium-binding domain-containing protein 3 | Rattus norvegicus (Rat) | PR |
| Q0JC44 | CML22 | Probable calcium-binding protein CML22 | Oryza sativa subsp japonica (Rice) | PR |
| Q9FYK2 | CML25 | Probable calcium-binding protein CML25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRN6 | CML22 | Probable calcium-binding protein CML22 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKRNGTRLNF | AKANSTKSGS | TRARDLMNKM | SIIPEWHSLY | SNTRKLPCSR | PHSSPCKMQR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TIFQDLCRPS | SSTTAIANPV | LSYLDIERIL | AQKISSRRDD | IKKVFQILDR | NHNQMVTKGD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LKRVITAFLI | PLTKDQFQDL | LAQIPISSLG | NVPYLEFLSR | FGGGIDININ | GIKRVNENEV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DNRRTVKEVQ | LTEKIFRNMR | SIRKVFQVMD | VNNTGLVQPQ | ELRRVLETFC | LRMQDGDYEK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FLEQYNIDKT | TAVDYNAFLK | NLSVKNDASF | KYLLSNAAEL | SRETQQGKNG | KRLLDTGSSE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DVWKNYSLDD | LEKTFCQEFS | KSYEKIEKAL | SAGDPSKGGY | ISLNYLKVVL | DTFIYRLPRR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| IFIQLIRRFG | LKTSTKINWK | QFLTAIYERQ | KLEVSKTLPL | KKRSSTEARN | RSRKENIIKK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LFKYSEDRYT | ALKKTLLIIS | STPSGHITWE | ELRHILNCMV | AKLNDSEFSE | LKQTFDPEGT |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GAVRVNSLLD | VLDDSTKVRK | MSPSTDTKTP | LPVAWDSVEE | LVLDSITRNL | QAFYSMLQSY |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DLRDTGTIGK | NNFRKVMRVF | CPYLSNEHLV | RFSSKFQEAG | SGRILYKKFL | LSIGVGIPPP |
| 610 | 620 | 630 | 640 | 650 | 660 |
| TPPLSSPKVQ | LSDQFQIEEP | GQQDERTQPS | GEKTSEINNM | TKEEVIDGLK | HRIQQKDPVF |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RKQFLSISKE | PDVKINQEEF | RKVLERSGMP | MNDCQYAMLA | SKLGFKNEEG | MSYQDFTMGF |
| 730 | 740 | 750 | 760 | 770 | 780 |
| EDCMLSGLET | VPLQSRTASR | TNMDEHFISA | EECLRIFPKK | LKESFRDVYS | AFFRIDLDRD |
| 790 | 800 | 810 | 820 | 830 | 840 |
| GIISMHDFHR | LLQYLQLNMV | DLEFERFLSL | LGLRLSVTLN | FREFQNLCEK | RPWKSDEAPQ |
| 850 | 860 | 870 | 880 | 890 | 900 |
| RLIRCKQKVA | DSELACEQAH | QYLIMKAKTR | WADLSKNFIE | TDNEGNGILR | RRDIKNSLYG |
| 910 | 920 | 930 | 940 | 950 | 960 |
| FDIPLTPREF | EKLWQNYDTE | GRGYITYQEF | LHRLGIRYSP | KVHRPYKEDY | FNFLGHFTKP |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| KQVQEEIQEL | QQISEREKLM | NHYEEISKAF | NAMEKSKPVA | LCRVQKVLQE | CGCPLKEEEL |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| ISLLKSLDVS | VHNNHIDPVE | FLRALEISWA | SKARPKEKEE | SSPPPISFSK | VTPDEVIKTM |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| QEVVESSQPA | LVEAFSALDK | EDTGFVKAME | FGDVLRSVCQ | KLTDNQYHYF | LRRLRLHLTP |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| NIHWKYFLEN | FSTFQDETAD | DWAENMPKAP | PPMSPKETAH | RDIVARVQKA | VASHYHTIVQ |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| EFENFDTLKS | NTVSRDEFRS | ICTRHIQILT | DEQFDRLWSE | LPVNAKGRLK | YQDFLSKLSI |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| ERVPSPPMAA | GDSGESTMAQ | RGSSAPEFSQ | GTRSNLYSPP | RDSRVGLKSR | SHPCTPVGTP |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| PLQNCEPIES | RLRKQIQGCW | RELLRECKEK | DTDKQGTISA | AEFLALVEKF | KLDISREESQ |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| QLIVKYDLKN | NGKFAYCDFI | QSCVLLLKAK | ETSLMRRMRI | QNADKMKEAG | METPSFYSAL |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| LRIQPKIVHC | WRPMRRSFKT | YDKNGTGLLS | VADFRKVLRQ | YSINLSEEEF | FHVLEYYDKS |
| 1510 | |||||
| LSSKISYNDF | LRAFLQ |