Q6GMN2
Gene name |
Baiap2 |
Protein name |
BAR/IMD domain-containing adapter protein 2 |
Names |
Brain-specific angiogenesis inhibitor 1-associated protein 2 , BAI-associated protein 2 , BAI1-associated protein 2 , Insulin receptor substrate protein of 53 kDa , IRSp53 , Insulin receptor substrate p53 , Insulin receptor tyrosine kinase substrate protein p53 |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:117542 |
EC number |
|
Protein Class |
|
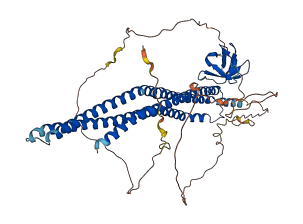
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
180-318 (Central region) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Krugmann S et al. (2001) "Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex", Current biology : CB, 11, 1645-55
- Miki H et al. (2002) "WAVE2 serves a functional partner of IRSp53 by regulating its interaction with Rac", Biochemical and biophysical research communications, 293, 93-9
- Kast DJ et al. (2014) "Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors", Nature structural & molecular biology, 21, 413-22
Autoinhibited structure

Activated structure

1 structures for Q6GMN2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6GMN2-F1 | Predicted | AlphaFoldDB |
No variants for Q6GMN2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q6GMN2 | |||||
No associated diseases with Q6GMN2
4 regional properties for Q6GMN2
Functions
24 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendritic shaft | Cylindric portion of the dendrite, directly stemming from the perikaryon, and carrying the dendritic spines. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| dendritic spine cytoplasm | The region of the neuronal cytoplasm located in dendritic spines. |
| excitatory synapse | A synapse in which an action potential in the presynaptic cell increases the probability of an action potential occurring in the postsynaptic cell. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| neuron projection branch point | The location where a secondary projection arises from a neuron projection. |
| neuron projection terminus | The specialized, terminal region of a neuron projection such as an axon or a dendrite. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| postsynapse | The part of a synapse that is part of the post-synaptic cell. |
| postsynaptic cytosol | The region of the cytosol consisting of all cytosol that is part of the postsynapse. |
| postsynaptic density, intracellular component | A network of proteins adjacent to the postsynaptic membrane forming an electron dense disc. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize neurotransmitter receptors in the adjacent membrane, such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
| presynaptic cytosol | The region of the cytosol consisting of all cytosol that is part of the presynapse. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
| Schaffer collateral - CA1 synapse | A synapse between the Schaffer collateral axon of a CA3 pyramidal cell and a CA1 pyramidal cell. |
| secretory granule | A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules. |
| synaptic membrane | A specialized area of membrane on either the presynaptic or the postsynaptic side of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| cytoskeletal anchor activity | The binding activity of a protein that brings together a cytoskeletal protein (either a microtubule or actin filament, spindle pole body, or protein directly bound to them) and one or more other molecules, permitting them to function in a coordinated way. |
| identical protein binding | Binding to an identical protein or proteins. |
| PDZ domain binding | Binding to a PDZ domain of a protein, a domain found in diverse signaling proteins. |
| proline-rich region binding | Binding to a proline-rich region, i.e. a region that contains a high proportion of proline residues, in a protein. |
| scaffold protein binding | Binding to a scaffold protein. Scaffold proteins are crucial regulators of many key signaling pathways. Although not strictly defined in function, they are known to interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. |
| transcription coregulator binding | Binding to a transcription coregulator, a protein involved in regulation of transcription via protein-protein interactions with transcription factors and other transcription regulatory proteins. Cofactors do not bind DNA directly, but rather mediate protein-protein interactions between regulatory transcription factors and the basal transcription machinery. |
18 GO annotations of biological process
| Name | Definition |
|---|---|
| actin crosslink formation | The process in which two or more actin filaments are connected together by proteins that act as crosslinks between the filaments. The crosslinked filaments may be on the same or differing axes. |
| actin filament bundle assembly | The assembly of actin filament bundles; actin filaments are on the same axis but may be oriented with the same or opposite polarities and may be packed with different levels of tightness. |
| cellular response to epidermal growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an epidermal growth factor stimulus. |
| cellular response to L-glutamate | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a L-glutamate(1-) stimulus. |
| dendrite development | The process whose specific outcome is the progression of the dendrite over time, from its formation to the mature structure. |
| modulation of chemical synaptic transmission | Any process that modulates the frequency or amplitude of synaptic transmission, the process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse. Amplitude, in this case, refers to the change in postsynaptic membrane potential due to a single instance of synaptic transmission. |
| neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| plasma membrane organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the plasma membrane. |
| positive regulation of actin filament polymerization | Any process that activates or increases the frequency, rate or extent of actin polymerization. |
| positive regulation of dendritic spine morphogenesis | Any process that increases the rate, frequency, or extent of dendritic spine morphogenesis, the process in which the anatomical structures of a dendritic spine are generated and organized. A dendritic spine is a protrusion from a dendrite and a specialized subcellular compartment involved in synaptic transmission. |
| positive regulation of excitatory postsynaptic potential | Any process that enhances the establishment or increases the extent of the excitatory postsynaptic potential (EPSP) which is a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. The flow of ions that causes an EPSP is an excitatory postsynaptic current (EPSC) and makes it easier for the neuron to fire an action potential. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| protein localization to synapse | Any process in which a protein is transported to, and/or maintained at the synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of modification of postsynaptic actin cytoskeleton | Any process that modulates the frequency, rate or extent of modification of postsynaptic actin cytoskeleton. |
| regulation of postsynapse organization | Any process that modulates the physical form of a postsynapse. |
| regulation of synaptic plasticity | A process that modulates synaptic plasticity, the ability of synapses to change as circumstances require. They may alter function, such as increasing or decreasing their sensitivity, or they may increase or decrease in actual numbers. |
3 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5EAD0 | BAIAP2 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | Bos taurus (Bovine) | PR |
| Q9UQB8 | BAIAP2 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | Homo sapiens (Human) | EV |
| Q8BKX1 | Baiap2 | BAR/IMD domain-containing adapter protein 2 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLSRSEEMH | RLTENVYKTI | MEQFNPSLRN | FIAMGKNYEK | ALAGVTFAAK | GYFDALVKMG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ELASESQGSK | ELGDVLFQMA | EVHRQIQNQL | EEMLKAFHNE | LLTQLEQKVE | LDSRYLSAAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KKYQTEQRSK | GDALDKCQAE | LKKLRKKSQG | SKNPQKYSDK | ELQYIDAISN | KQGELENYVS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DGYKTALTEE | RRRFCFLVEK | QCAVAKNSAA | YHSKGKELLA | QKLPLWQQAC | ADPNKIPDRA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AQLMQQMANS | NGSILPSALS | ASKSNLVISD | PIPGAKPLPV | PPELAPFVGR | MSAQENVPVM |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NGVAGADSED | YNPWADRKAA | QPKSLSPPQS | QSKLSDSYSN | TLPVRKSVTP | KNSYATTENK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TLPRSSSMAA | GLERNGRMRV | KAIFSHAAGD | NSTLLSFKEG | DLITLLVPEA | RDGWHYGESE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KTKMRGWFPF | SYTRVLDSDG | SDRLHMSLQQ | GKSSSTGNLL | DKDDLALPPP | DYGTSSRAFP |
| 490 | 500 | 510 | 520 | 530 | |
| SQTAGTFKQR | PYSVAVPAFS | QGLDDYGARS | VSRNPFANVH | LKPTVTNDRS | APLLS |