Q6DN90
Gene name |
IQSEC1 (ARFGEP100, BRAG2, KIAA0763) |
Protein name |
IQ motif and SEC7 domain-containing protein 1 |
Names |
ADP-ribosylation factors guanine nucleotide-exchange protein 100 , ADP-ribosylation factors guanine nucleotide-exchange protein 2 , Brefeldin-resistant Arf-GEF 2 protein , BRAG2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9922 |
EC number |
|
Protein Class |
|
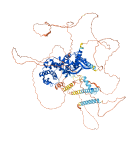
Descriptions
IQ motif and SEC7 domain-containing protein 1 (IQSEC1, also known as BRAG2) is a guanine nucleotide exchange factor (GEF) that is highly enriched in synapses. The autoinhibition of IQSEC1 is mediated by its IQ motif, which binds to apo-CaM and folds together with its Sec7-PH tandem, leading IQSEC1 to adopt a closed conformation in the absence of Ca2+. This autoinhibition is released by calcium binding, which triggers a conformational change that exposes the DH domain for activation.
Autoinhibitory domains (AIDs)
Target domain |
517-864 (Sec7-PH domains) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1049 variants for Q6DN90
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001263384 rs751369634 COSM3914893 COSM3914895 |
55 | P>L | Intellectual disability Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
NCI-TCGA Cosmic ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
rs769383880 RCV001331976 |
277 | R>Q | Intellectual developmental disorder with short stature and behavioral abnormalities [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001331977 rs1698161896 |
331 | D>Y | Intellectual developmental disorder with short stature and behavioral abnormalities [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV000860024 rs758170522 VAR_083480 |
335 | R>Q | Intellectual developmental disorder with short stature and behavioral abnormalities IDDSSBA; loss of function [ClinVar, UniProt] | Yes |
ClinVar UniProt ExAC TOPMed dbSNP gnomAD |
|
RCV000860023 VAR_083481 rs765723607 |
357 | T>M | Intellectual developmental disorder with short stature and behavioral abnormalities IDDSSBA; loss of function [ClinVar, UniProt] | Yes |
ClinVar UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs2125307036 RCV001843720 |
402 | K>missing | Developmental disorder [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000885046 rs17541405 RCV002495359 VAR_027004 |
882 | V>I | Intellectual developmental disorder with short stature and behavioral abnormalities [ClinVar] | Yes |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1553666557 | 2 | W>* | No | Ensembl | |
| rs1700653924 | 2 | W>* | No | TOPMed | |
| rs1576071511 | 2 | W>R | No | Ensembl | |
| rs1180063283 | 3 | C>Y | No | gnomAD | |
| rs1476336906 | 4 | L>P | No |
TOPMed gnomAD |
|
| rs1700653030 | 5 | H>R | No | TOPMed | |
| rs1196677361 | 6 | C>G | No | TOPMed | |
| rs772658798 | 7 | N>D | No |
ExAC gnomAD |
|
| rs1700652314 | 7 | N>T | No | Ensembl | |
| rs1700651956 | 8 | S>* | No | TOPMed | |
| rs1700652126 | 8 | S>A | No |
TOPMed gnomAD |
|
| rs1700651413 | 10 | R>G | No | Ensembl | |
| rs1210587898 | 10 | R>K | No | gnomAD | |
| rs1481052567 | 12 | Q>* | No | TOPMed | |
| rs1158875511 | 12 | Q>R | No | TOPMed | |
| rs1700650321 | 13 | S>P | No | TOPMed | |
| rs1329743207 | 14 | L>I | No | gnomAD | |
| rs1054487734 | 16 | E>Q | No |
TOPMed gnomAD |
|
| rs926747306 | 20 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1447783148 | 22 | G>S | No |
TOPMed gnomAD |
|
| rs754774497 | 23 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs766330520 | 24 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1204515649 | 25 | G>D | No | gnomAD | |
| rs1232204506 | 25 | G>S | No | Ensembl | |
| rs142851488 | 26 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1698780740 | 27 | A>T | No | Ensembl | |
| rs1229605668 | 29 | S>I | No | gnomAD | |
| rs1341826870 | 30 | S>N | No | gnomAD | |
| rs1221473553 | 31 | E>G | No | gnomAD | |
|
COSM123775 rs1295853946 |
31 | E>K | upper_aerodigestive_tract [Cosmic] | No |
cosmic curated gnomAD |
| rs886125872 | 32 | T>I | No | Ensembl | |
| rs865993734 | 33 | G>A | No |
TOPMed gnomAD |
|
| rs865993734 | 33 | G>D | No |
TOPMed gnomAD |
|
|
COSM6096271 rs865993734 COSM581401 |
33 | G>V | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic TOPMed gnomAD |
| rs898001891 | 34 | T>A | No |
TOPMed gnomAD |
|
| rs1698778373 | 34 | T>I | No | TOPMed | |
| COSM4826742 | 35 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1372652424 | 36 | L>P | No | gnomAD | |
| TCGA novel | 37 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1308807967 | 37 | D>N | No | gnomAD | |
| rs375594309 | 39 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs375594309 | 39 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1407433890 | 40 | S>* | No | gnomAD | |
| rs1407433890 | 40 | S>L | No | gnomAD | |
| rs771331757 | 43 | P>S | No |
ExAC gnomAD |
|
| rs749700860 | 46 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs749700860 | 46 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1476716670 | 48 | V>M | No | gnomAD | |
| rs139752173 | 50 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs755217989 | 54 | S>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 54 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2125356177 | 56 | D>Y | No | Ensembl | |
| rs758231467 | 57 | H>D | No |
ExAC gnomAD |
|
| rs765197194 | 57 | H>Q | No |
ExAC gnomAD |
|
| COSM4896109 | 57 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs866633619 | 58 | Y>H | No | Ensembl | |
| rs753404458 | 59 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs1698771527 | 59 | E>G | No | TOPMed | |
| rs753404458 | 59 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1698771138 | 60 | H>R | No | gnomAD | |
| rs1409483537 | 60 | H>Y | No | gnomAD | |
| rs753528211 | 61 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1359674838 | 62 | S>L | No | gnomAD | |
| rs1467350927 | 62 | S>T | No | gnomAD | |
|
rs376777987 COSM365233 |
63 | V>L | lung [Cosmic] | No |
cosmic curated ESP TOPMed gnomAD |
| rs1467785898 | 64 | G>R | No |
TOPMed gnomAD |
|
| rs1191184868 | 65 | A>T | No | TOPMed | |
| rs933329955 | 65 | A>V | No |
TOPMed gnomAD |
|
| rs150511898 | 66 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1177071408 | 67 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs148012608 | 68 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1483751664 | 69 | Y>C | No | gnomAD | |
| rs1183972314 | 69 | Y>H | No | gnomAD | |
| rs1349493270 | 70 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1349493270 | 70 | S>W | No |
TOPMed gnomAD |
|
| rs1328003505 | 72 | P>L | No | Ensembl | |
| TCGA novel | 72 | P>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780351811 | 73 | P>A | No |
ExAC gnomAD |
|
| rs758094163 | 73 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs542614009 | 74 | G>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs753778368 | 75 | Q>K | No |
ExAC gnomAD |
|
| rs763456049 | 76 | Q>* | No |
ExAC gnomAD |
|
| TCGA novel | 76 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760255548 | 76 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1450423723 | 77 | Q>H | No | gnomAD | |
| rs752254359 | 78 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs767241790 | 78 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs767241790 | 78 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs763391103 | 79 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs763391103 | 79 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs763391103 | 79 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs142179028 | 80 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs777214549 | 80 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs142179028 | 80 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs768575504 | 81 | R>G | No |
ExAC gnomAD |
|
| rs1698761782 | 83 | K>R | No | TOPMed | |
| rs1698761385 | 85 | Q>R | No | gnomAD | |
| rs747150592 | 86 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1698760921 | 86 | H>R | No | TOPMed | |
| rs556945444 | 87 | S>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs556945444 | 87 | S>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1575991344 | 89 | S>P | No | Ensembl | |
| rs1395522466 | 90 | I>F | No | TOPMed | |
| rs778611987 | 90 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs1395522466 | 90 | I>V | No | TOPMed | |
| rs777798451 | 92 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs755549936 | 92 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs755549936 | 92 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs777798451 | 92 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1698757690 | 93 | K>Q | No | TOPMed | |
| TCGA novel | 93 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1698757464 | 94 | Q>H | No | gnomAD | |
| rs752303829 | 95 | A>T | No |
ExAC gnomAD |
|
| rs1226144922 | 96 | E>G | No | gnomAD | |
| rs1698756829 | 96 | E>K | No | TOPMed | |
| rs1294739176 | 97 | E>* | No |
TOPMed gnomAD |
|
| rs754747708 | 97 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1294739176 | 97 | E>Q | No |
TOPMed gnomAD |
|
| rs1444367293 | 99 | A>V | No | TOPMed | |
| rs751266418 | 100 | I>L | No |
ExAC gnomAD |
|
| rs765643763 | 102 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM4113446 COSM4113444 rs536974018 |
102 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs536974018 | 102 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1698754027 | 104 | R>G | No | Ensembl | |
| rs1054317672 | 104 | R>H | No |
TOPMed gnomAD |
|
| rs575906745 | 108 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs746031903 | 109 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1698751818 | 110 | Y>C | No |
TOPMed gnomAD |
|
| rs774578154 | 112 | L>F | No |
ExAC gnomAD |
|
| rs1227619909 | 112 | L>P | No | gnomAD | |
| rs867431824 | 114 | S>* | No | gnomAD | |
| rs867431824 | 114 | S>L | No | gnomAD | |
| COSM3914891 | 116 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1698750021 | 117 | Q>* | No |
TOPMed gnomAD |
|
| rs995060076 | 118 | D>N | No | gnomAD | |
| rs1698749458 | 120 | Q>K | No |
TOPMed gnomAD |
|
| rs1575971472 | 121 | V>G | No | Ensembl | |
| rs1257998149 | 121 | V>M | No | gnomAD | |
| rs1559646128 | 123 | M>I | No | Ensembl | |
| rs1201035554 | 126 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs1322692577 COSM4113442 |
126 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs901714702 | 128 | Y>H | No |
TOPMed gnomAD |
|
| rs1329433630 | 131 | R>C | No | gnomAD | |
| rs759644080 | 131 | R>H | No | ExAC | |
| rs1575971324 | 132 | L>R | No | Ensembl | |
| rs1287256576 | 133 | V>I | No |
TOPMed gnomAD |
|
| rs1287256576 | 133 | V>L | No |
TOPMed gnomAD |
|
| rs1037079317 | 134 | T>P | No | Ensembl | |
| rs774526868 | 135 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs774526868 | 135 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1698230640 | 135 | R>H | No | Ensembl | |
| rs1342703452 | 136 | H>R | No | gnomAD | |
| TCGA novel | 138 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762645084 | 139 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs762645084 | 139 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM1418755 rs1161508071 |
139 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs149221542 | 140 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1411527644 | 141 | I>L | No |
TOPMed gnomAD |
|
| rs769485180 | 143 | T>M | No |
ExAC gnomAD |
|
|
COSM1210867 rs781305360 |
144 | A>V | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1698227069 | 145 | F>L | No | TOPMed | |
| COSM3914889 | 146 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1698226833 | 146 | R>H | No | TOPMed | |
| rs1698226833 | 146 | R>L | No | TOPMed | |
| TCGA novel | 147 | Q>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 149 | Q>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1575971087 | 150 | M>L | No | Ensembl | |
| rs1210069273 | 154 | F>L | No |
TOPMed gnomAD |
|
| rs1282934572 | 156 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1698225322 | 156 | R>H | No | Ensembl | |
|
rs549147535 COSM1038158 |
158 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
|
COSM4619800 COSM4619802 rs1285176495 |
158 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
rs1285176495 COSM4138433 |
158 | R>L | ovary [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1244463782 | 159 | S>T | No | gnomAD | |
| rs778140784 | 160 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs372619629 | 161 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1316276894 | 163 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1414741648 | 165 | R>H | No | gnomAD | |
| rs753209268 | 166 | M>R | No |
ExAC gnomAD |
|
| rs753209268 | 166 | M>T | No |
ExAC gnomAD |
|
| rs1698221817 | 168 | R>C | No | Ensembl | |
| rs768053121 | 168 | R>H | No |
ExAC gnomAD |
|
| rs759591010 | 169 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs759591010 | 169 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1698221364 | 169 | R>W | No | Ensembl | |
| rs1575970841 | 171 | V>L | No | Ensembl | |
| rs766586796 | 175 | M>L | No |
ExAC gnomAD |
|
| rs763171910 | 175 | M>T | No |
ExAC gnomAD |
|
| rs766586796 | 175 | M>V | No |
ExAC gnomAD |
|
| rs1559645599 | 176 | R>G | No | Ensembl | |
| rs1488173273 | 179 | F>L | No | TOPMed | |
| rs1265105992 | 183 | G>R | No | TOPMed | |
| rs1453000724 | 187 | V>L | No |
TOPMed gnomAD |
|
| rs1453000724 | 187 | V>M | No |
TOPMed gnomAD |
|
| rs1290070371 | 188 | H>Y | No | gnomAD | |
| rs1222240212 | 189 | S>N | No | gnomAD | |
| rs1222240212 | 189 | S>T | No | gnomAD | |
| rs1357819581 | 190 | S>P | No | gnomAD | |
| rs527631233 | 191 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1379438887 | 192 | F>I | No | TOPMed | |
| rs761549809 | 192 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1379438887 | 192 | F>V | No | TOPMed | |
| rs989850235 | 193 | E>K | No |
TOPMed gnomAD |
|
| rs776684248 | 194 | G>R | No |
ExAC gnomAD |
|
| rs1698215821 | 195 | K>E | No | Ensembl | |
| rs1438655656 | 196 | Q>E | No | gnomAD | |
| rs1350698755 | 198 | S>L | No | gnomAD | |
| rs1698214932 | 199 | V>G | No | TOPMed | |
| rs746511594 | 201 | N>D | No |
ExAC gnomAD |
|
| rs575762365 | 202 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 202 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs771494706 | 202 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs778461158 | 203 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs778461158 | 203 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1698211725 | 204 | S>P | No | TOPMed | |
| rs756471479 | 205 | Q>H | No |
ExAC gnomAD |
|
| rs1698211124 | 206 | L>P | No | Ensembl | |
| rs368630864 | 208 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1375113842 | 210 | V>L | No | TOPMed | |
| rs1698209420 | 211 | S>F | No | TOPMed | |
| rs2125312077 | 211 | S>P | No | Ensembl | |
| rs752069173 | 212 | P>L | No |
ExAC gnomAD |
|
| rs752069173 | 212 | P>R | No |
ExAC gnomAD |
|
| rs755548341 | 212 | P>S | No |
ExAC gnomAD |
|
| rs1242629609 | 213 | E>G | No |
TOPMed gnomAD |
|
| rs1698206825 | 214 | C>* | No | Ensembl | |
| rs766428334 | 214 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1698206573 | 215 | G>S | No | Ensembl | |
| rs1575970332 | 216 | D>A | No | Ensembl | |
| rs750623715 | 218 | S>I | No |
ExAC gnomAD |
|
| rs1321212413 | 218 | S>R | No |
TOPMed gnomAD |
|
| rs1575970270 | 219 | E>G | No | Ensembl | |
|
COSM4583926 rs200588747 COSM1038157 |
219 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1309650381 | 220 | P>R | No |
TOPMed gnomAD |
|
| rs1698203884 | 221 | T>I | No | TOPMed | |
| rs1457617498 | 222 | T>I | No |
TOPMed gnomAD |
|
| rs1575970194 | 222 | T>P | No | Ensembl | |
| rs1416100544 | 224 | K>M | No | gnomAD | |
| TCGA novel | 224 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745410560 | 225 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs745410560 | 225 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs771570751 | 225 | S>P | No |
ExAC gnomAD |
|
| rs745410560 | 225 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs374637598 | 226 | P>L | No |
ESP TOPMed gnomAD |
|
| rs1237374778 | 226 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1698201200 | 227 | A>S | No | TOPMed | |
| rs770548389 | 228 | P>L | No |
ExAC gnomAD |
|
| rs1480543701 | 229 | S>C | No | gnomAD | |
| rs1212908009 | 230 | S>C | No | gnomAD | |
| rs1422871641 | 230 | S>N | No | Ensembl | |
| rs748509845 | 232 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs781471784 | 233 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs749302895 | 233 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs142197279 | 235 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1698198196 | 235 | A>V | No | Ensembl | |
| rs368428333 | 238 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1575969908 | 241 | D>A | No | Ensembl | |
|
rs754101583 COSM1418753 |
242 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1698196318 | 243 | F>L | No | Ensembl | |
| rs1332306971 | 245 | R>G | No |
TOPMed gnomAD |
|
| rs1335537073 | 245 | R>S | No | gnomAD | |
| rs143857704 | 247 | V>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs771082004 | 247 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs775599935 | 251 | A>S | No |
ExAC gnomAD |
|
| rs759098618 | 252 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1322362814 | 254 | I>T | No |
TOPMed gnomAD |
|
| rs1183912558 | 254 | I>V | No | gnomAD | |
|
TCGA novel rs1698193511 |
255 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
rs770468770 COSM1669982 |
256 | D>N | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs748994453 | 257 | A>S | No |
ExAC gnomAD |
|
| rs1006242154 | 259 | N>K | No | TOPMed | |
| rs889088774 | 260 | C>* | No |
TOPMed gnomAD |
|
| rs769044871 | 261 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs747329812 | 261 | R>H | No |
ExAC gnomAD |
|
| rs1233227066 | 264 | H>R | No |
TOPMed gnomAD |
|
| rs780627819 | 264 | H>Y | No |
ExAC gnomAD |
|
| rs746178150 | 265 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1698189601 | 265 | T>I | No | Ensembl | |
| COSM4853952 | 266 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746385357 | 268 | A>S | No |
ExAC gnomAD |
|
| rs746385357 | 268 | A>T | No |
ExAC gnomAD |
|
|
rs201771792 COSM3060617 |
269 | P>L | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs754154698 | 270 | A>T | No |
ExAC gnomAD |
|
| rs1698187880 | 271 | L>V | No | Ensembl | |
| rs1698186995 | 272 | D>E | No | Ensembl | |
| rs1698187245 | 272 | D>G | No | gnomAD | |
| rs764372516 | 272 | D>H | No |
ExAC gnomAD |
|
| rs764372516 | 272 | D>N | No |
ExAC gnomAD |
|
| rs370578211 | 273 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM1418752 | 274 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs376868528 | 274 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs751498657 | 275 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs765919597 | 275 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs765919597 | 275 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs751498657 | 275 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs769383880 | 277 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs772878747 | 277 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs755430529 | 278 | D>N | No | Ensembl | |
| rs761035991 | 279 | T>I | No |
ExAC gnomAD |
|
| rs1314069234 | 280 | E>D | No | gnomAD | |
| rs570967760 | 280 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM4113434 | 280 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746413127 | 281 | P>S | No |
ExAC gnomAD |
|
| COSM3846180 | 282 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1386366332 | 282 | Q>R | No |
TOPMed gnomAD |
|
|
rs143647610 RCV000968559 |
283 | T>I | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs143647610 | 283 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs749332755 | 284 | A>G | No |
ExAC gnomAD |
|
| rs771166621 | 284 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs771166621 | 284 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs756336349 | 287 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs139139699 | 287 | G>D | No |
ESP ExAC gnomAD |
|
| rs756336349 | 287 | G>R | No |
ExAC TOPMed gnomAD |
|
|
COSM419837 rs756336349 |
287 | G>S | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs199808138 | 288 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751488740 | 291 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs114114109 | 291 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs111660488 | 292 | K>R | No |
TOPMed gnomAD |
|
| rs111660488 | 292 | K>T | No |
TOPMed gnomAD |
|
| rs749873023 | 293 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs370536991 | 294 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1430554083 | 295 | E>K | No |
TOPMed gnomAD |
|
| rs1698176880 | 297 | T>A | No | TOPMed | |
| rs761433310 | 297 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1344228807 | 298 | A>D | No | gnomAD | |
| rs776326543 | 299 | S>L | No |
ExAC gnomAD |
|
| rs759905989 | 301 | S>G | No |
ExAC gnomAD |
|
| rs775020933 | 302 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1698174776 | 303 | V>G | No | TOPMed | |
| rs771507649 | 303 | V>I | No |
ExAC gnomAD |
|
| rs1344428078 | 304 | T>I | No | gnomAD | |
| rs1698173821 | 305 | L>P | No | TOPMed | |
| rs1376363406 | 308 | D>E | No | TOPMed | |
| rs769979846 | 308 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs984910197 | 310 | E>K | No |
TOPMed gnomAD |
|
| rs868368530 | 312 | L>V | No | Ensembl | |
|
rs1342413854 COSM4879002 |
313 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1698170874 | 314 | P>S | No |
TOPMed gnomAD |
|
|
COSM3060607 COSM3060605 |
315 | P>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1698170377 | 315 | P>R | No | gnomAD | |
| rs781442574 | 315 | P>S | No |
ExAC gnomAD |
|
| rs1575968690 | 317 | P>T | No | Ensembl | |
| rs1698168963 | 318 | L>V | No | Ensembl | |
| rs139140830 | 319 | S>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1698168376 | 319 | S>P | No |
TOPMed gnomAD |
|
| rs1482472747 | 320 | Q>R | No |
TOPMed gnomAD |
|
| rs1698166638 | 321 | A>T | No | Ensembl | |
| rs1698165831 | 322 | G>E | No | gnomAD | |
| rs758400333 | 322 | G>R | No |
ExAC gnomAD |
|
| TCGA novel | 323 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750350018 | 323 | D>N | No |
ExAC gnomAD |
|
| rs748366559 | 324 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs748366559 | 324 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs764778594 | 324 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs561571211 | 325 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1246567357 | 326 | S>T | No | TOPMed | |
| rs759831591 | 328 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1021596365 | 328 | T>I | No |
TOPMed gnomAD |
|
| rs1021596365 | 328 | T>S | No |
TOPMed gnomAD |
|
| rs1157035864 | 329 | E>D | No |
TOPMed gnomAD |
|
| rs766844830 | 329 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1328575200 | 330 | S>L | No |
TOPMed gnomAD |
|
| rs1698161896 | 331 | D>H | No | Ensembl | |
| rs748215013 | 333 | R>P | No |
ExAC TOPMed gnomAD |
|
|
COSM4113431 COSM4113429 rs748215013 |
333 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs773831156 | 333 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs776736809 | 334 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs776736809 | 334 | L>Q | No |
ExAC TOPMed gnomAD |
|
| rs758170522 | 335 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs747195858 | 335 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1559643838 | 337 | G>R | No | Ensembl | |
| rs372228887 | 339 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1203770698 | 339 | A>V | No | gnomAD | |
| rs1260703450 | 341 | P>A | No |
TOPMed gnomAD |
|
| rs372618665 | 341 | P>L | No | Ensembl | |
| rs1260703450 | 341 | P>S | No |
TOPMed gnomAD |
|
| rs1350878232 | 343 | Y>C | No |
TOPMed gnomAD |
|
| rs1350878232 | 343 | Y>F | No |
TOPMed gnomAD |
|
| rs1698156885 | 343 | Y>H | No | Ensembl | |
| rs1237251304 | 344 | W>* | No | gnomAD | |
| TCGA novel | 344 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763657553 | 344 | W>R | No |
ExAC gnomAD |
|
| rs755862339 | 345 | A>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 346 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1205274160 | 346 | L>P | No | TOPMed | |
| rs1316418820 | 348 | H>N | No |
TOPMed gnomAD |
|
| rs1316418820 | 348 | H>Y | No |
TOPMed gnomAD |
|
| rs752382828 | 349 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1404049674 | 350 | E>Q | No | gnomAD | |
| rs1322618825 | 351 | D>Y | No |
TOPMed gnomAD |
|
| rs1698154214 | 352 | K>R | No |
TOPMed gnomAD |
|
| rs1384481760 | 353 | A>D | No |
TOPMed gnomAD |
|
| rs1698153150 | 354 | D>N | No | TOPMed | |
| rs1306290858 | 355 | T>A | No |
TOPMed gnomAD |
|
|
COSM1038156 rs763376715 |
355 | T>M | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs765723607 | 357 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs765723607 | 357 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1698150593 | 358 | S>C | No | Ensembl | |
| rs776840677 | 358 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1162804503 | 359 | C>R | No |
TOPMed gnomAD |
|
| rs768608622 | 360 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs202190518 | 360 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs768608622 | 360 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs775661699 | 362 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs745615824 | 363 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs374393523 | 364 | S>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1698147152 | 366 | E>K | No | gnomAD | |
| rs755555955 | 367 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs777397340 | 367 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs148427042 | 369 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs79617847 | 369 | E>G | No |
ExAC gnomAD |
|
| rs1438081657 | 370 | Q>R | No | Ensembl | |
| rs750755282 | 371 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1367258246 | 371 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs754280926 | 373 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs754280926 | 373 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs762444448 | 373 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1004712778 | 374 | V>G | No | gnomAD | |
| rs764277311 | 374 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1190624660 | 375 | E>G | No | gnomAD | |
| rs181320071 | 376 | H>Y | No |
1000Genomes ExAC gnomAD |
|
| rs1032837409 | 378 | P>L | No | TOPMed | |
| rs1698140384 | 379 | L>P | No | Ensembl | |
| rs1357936197 | 380 | L>F | No | gnomAD | |
| rs1698139671 | 381 | T>A | No | Ensembl | |
| TCGA novel | 381 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1418572318 | 382 | I>L | No |
TOPMed gnomAD |
|
| rs1275658107 | 382 | I>M | No |
TOPMed gnomAD |
|
|
COSM1616875 rs1418572318 |
382 | I>V | liver [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs770619488 | 383 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1338899924 | 384 | P>Q | No | gnomAD | |
| rs749294733 | 385 | P>S | No |
ExAC gnomAD |
|
| rs138404122 | 386 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1346343486 | 387 | D>N | No |
TOPMed gnomAD |
|
| rs1303537602 | 390 | V>A | No |
TOPMed gnomAD |
|
| rs769457169 | 391 | D>N | No |
ExAC gnomAD |
|
| rs368148999 | 391 | D>V | No |
ESP TOPMed |
|
| rs977449124 | 392 | L>V | No | Ensembl | |
| rs781000761 | 393 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1698135294 | 394 | D>E | No | TOPMed | |
| rs754606659 | 395 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs866307730 | 395 | R>H | No |
TOPMed gnomAD |
|
| rs866307730 | 395 | R>L | No |
TOPMed gnomAD |
|
|
rs751161861 COSM1418751 COSM4113426 |
396 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1186202 COSM4113424 rs899865181 |
398 | R>Q | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs199881804 | 398 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1698133029 | 399 | G>R | No | TOPMed | |
| rs202078774 | 401 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1234956995 | 402 | K>R | No | gnomAD | |
| rs1437736659 | 405 | S>G | No | gnomAD | |
| rs1271000302 | 405 | S>N | No | gnomAD | |
| rs1324550251 | 406 | A>V | No | gnomAD | |
| rs764461456 | 407 | Y>H | No |
ExAC gnomAD |
|
| rs764461456 | 407 | Y>N | No |
ExAC gnomAD |
|
| rs146656961 | 408 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150386585 | 408 | E>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs752707238 | 408 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs752707238 | 408 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs774414321 | 409 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs201031929 | 409 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1171641571 | 410 | S>G | No | TOPMed | |
| rs139859163 | 410 | S>R | No |
ESP TOPMed gnomAD |
|
| rs1698127335 | 411 | L>V | No | Ensembl | |
| TCGA novel | 412 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1286519504 | 412 | G>C | No |
TOPMed gnomAD |
|
| rs867530038 | 412 | G>D | No | Ensembl | |
| rs1286519504 | 412 | G>R | No |
TOPMed gnomAD |
|
| rs773220855 | 413 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1410683603 | 415 | Q>P | No | gnomAD | |
| rs747662513 | 416 | G>D | No |
ExAC gnomAD |
|
| rs111322824 | 419 | K>R | No | Ensembl | |
| rs1231610693 | 421 | G>D | No | gnomAD | |
| TCGA novel | 422 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776342464 | 424 | S>G | No |
ExAC gnomAD |
|
|
rs746716911 COSM6163227 COSM6163229 COSM1536463 |
425 | G>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs746716911 | 425 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs757663574 | 426 | A>S | No |
ExAC gnomAD |
|
| rs757663574 | 426 | A>T | No |
ExAC gnomAD |
|
| rs749559790 | 427 | P>S | No |
ExAC gnomAD |
|
| rs1224068829 | 428 | K>Q | No | gnomAD | |
| rs1372546841 | 428 | K>R | No |
TOPMed gnomAD |
|
| rs778257947 | 429 | S>C | No |
ExAC gnomAD |
|
| rs1698119315 | 429 | S>T | No | TOPMed | |
| rs1412713726 | 430 | L>F | No | gnomAD | |
| rs1698118720 | 430 | L>P | No | Ensembl | |
| rs753189267 | 431 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs753189267 | 431 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs71306027 | 432 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs565538660 | 432 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs565538660 | 432 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs71306027 | 432 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs548917993 | 433 | E>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs762818593 | 433 | E>K | No |
ExAC gnomAD |
|
| TCGA novel | 433 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs548917993 | 433 | E>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1418918870 | 434 | E>D | No |
TOPMed gnomAD |
|
| rs1698116114 | 434 | E>K | No | gnomAD | |
| rs755004691 | 435 | P>A | No |
TOPMed gnomAD |
|
| rs755004691 | 435 | P>S | No |
TOPMed gnomAD |
|
| rs755004691 | 435 | P>T | No |
TOPMed gnomAD |
|
| rs1698115162 | 436 | E>D | No | TOPMed | |
| rs142922898 | 438 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs142922898 | 438 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4802584 rs535113723 |
438 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1698113507 | 439 | P>H | No | Ensembl | |
| rs1698113507 | 439 | P>R | No | Ensembl | |
| rs766829615 | 439 | P>S | No | gnomAD | |
| rs756600213 | 440 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs756600213 | 440 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs756600213 | 440 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs551318496 | 440 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs775222518 | 441 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs775222518 | 441 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs957542214 | 441 | P>S | No |
TOPMed gnomAD |
|
| rs749970308 | 442 | P>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 443 | R>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs144790333 | 443 | R>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs979920002 | 444 | P>L | No |
TOPMed gnomAD |
|
| TCGA novel | 444 | P>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1698110781 | 444 | P>S | No | TOPMed | |
| rs1698110781 | 444 | P>T | No | TOPMed | |
| rs1645514565 | 445 | L>P | No | gnomAD | |
| rs369999833 | 446 | D>N | No | Ensembl | |
| rs756448075 | 451 | I>V | No |
ExAC gnomAD |
|
| rs914654971 | 452 | N>S | No | TOPMed | |
| COSM4113421 | 453 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1313779626 | 453 | G>S | No | gnomAD | |
| TCGA novel | 454 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755104723 | 455 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs781634093 | 455 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs755104723 | 455 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs118154115 | 457 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1359077143 | 457 | R>W | No |
TOPMed gnomAD |
|
| rs141042552 | 458 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1698106192 | 459 | S>G | No | Ensembl | |
| rs1385082698 | 459 | S>R | No | gnomAD | |
| rs1575965971 | 463 | S>A | No | Ensembl | |
| rs1430439098 | 463 | S>L | No |
TOPMed gnomAD |
|
| rs1485005812 | 464 | D>E | No | gnomAD | |
| rs1186192832 | 464 | D>G | No | gnomAD | |
| rs765167842 | 465 | Y>C | No |
ExAC gnomAD |
|
| rs1698103307 | 468 | G>D | No | TOPMed | |
| rs190016439 | 468 | G>S | No |
1000Genomes ExAC gnomAD |
|
|
COSM4812962 COSM445565 |
469 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1698102554 | 470 | N>S | No | Ensembl | |
| rs1242507635 | 471 | D>E | No | gnomAD | |
| rs760239151 | 473 | I>F | No |
ExAC gnomAD |
|
| rs150863500 | 473 | I>S | No |
ESP TOPMed gnomAD |
|
| rs760239151 | 473 | I>V | No |
ExAC gnomAD |
|
| rs1698101113 | 475 | S>G | No | TOPMed | |
| rs771599178 | 475 | S>T | No |
ExAC gnomAD |
|
| rs759160338 | 476 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1365328504 | 477 | S>F | No | gnomAD | |
| rs1331088355 | 477 | S>T | No |
TOPMed gnomAD |
|
| rs1162047634 | 478 | N>S | No | gnomAD | |
| rs1698099166 | 479 | S>C | No | Ensembl | |
|
rs1169772507 COSM4113419 |
481 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1373772542 | 481 | D>H | No | gnomAD | |
| rs1373772542 | 481 | D>N | No | gnomAD | |
| rs1698098183 | 482 | T>A | No | Ensembl | |
| rs148022111 | 483 | I>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1698097583 | 483 | I>V | No | Ensembl | |
| rs1698096773 | 486 | S>R | No | gnomAD | |
| rs1698096184 | 487 | S>F | No | Ensembl | |
| rs747542479 | 488 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1698095394 | 489 | S>T | No | TOPMed | |
| rs758477143 | 490 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1698094806 | 490 | S>P | No | TOPMed | |
| rs775312948 | 492 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs375728760 COSM4113417 |
492 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1559641967 | 493 | D>E | No | Ensembl | |
| rs760589697 | 496 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs760589697 | 496 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs766974740 | 497 | E>Q | No |
ExAC gnomAD |
|
| rs770512758 | 499 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs759043751 | 499 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs770512758 | 499 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs1166529097 | 500 | L>F | No | gnomAD | |
| COSM1038154 | 500 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2125304215 | 502 | K>M | No | Ensembl | |
| rs1698089757 | 503 | Q>P | No | Ensembl | |
| rs777045414 | 504 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs777045414 | 504 | T>P | No |
ExAC TOPMed gnomAD |
|
|
rs2125304158 RCV001806991 |
505 | Y>missing | No |
ClinVar dbSNP |
|
| TCGA novel | 506 | H>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747513020 | 510 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs747513020 | 510 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs780572634 COSM1038153 |
510 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs780572634 | 510 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1698087859 | 511 | N>D | No | Ensembl | |
| rs144594173 | 511 | N>K | No |
ESP gnomAD |
|
| rs779283641 | 520 | N>S | No |
ExAC gnomAD |
|
| rs1698086114 | 521 | D>G | No | TOPMed | |
| rs1456118893 | 521 | D>N | No | gnomAD | |
| rs1398520976 | 522 | V>I | No |
TOPMed gnomAD |
|
| rs1298913747 | 524 | R>C | No |
TOPMed gnomAD |
|
| rs141003089 | 524 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1698084174 | 528 | Y>* | No | Ensembl | |
| rs1698084401 | 528 | Y>C | No | Ensembl | |
|
COSM3060563 rs755905106 |
529 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs752605932 | 530 | I>V | No |
ExAC gnomAD |
|
| rs751000548 | 531 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs751000548 | 531 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1698082886 | 534 | L>V | No | TOPMed | |
| rs766240880 | 535 | F>L | No | Ensembl | |
| rs1157778151 | 538 | K>N | No |
TOPMed gnomAD |
|
| rs1035749368 | 543 | V>I | No | Ensembl | |
| rs1575925176 | 545 | Y>S | No | Ensembl | |
| rs1472213380 | 548 | E>K | No |
TOPMed gnomAD |
|
| rs1472213380 | 548 | E>Q | No |
TOPMed gnomAD |
|
|
COSM1327893 rs200196417 COSM4692927 |
549 | R>H | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs960829899 | 551 | F>S | No | TOPMed | |
| rs1696958033 | 553 | P>L | No | TOPMed | |
| rs1696958274 | 553 | P>S | No | TOPMed | |
| rs1696956935 | 554 | D>E | No | TOPMed | |
| rs750067475 | 554 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs764890738 | 555 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1362238013 | 556 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs377447856 | 557 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs377447856 | 557 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1049023791 | 558 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
COSM6048209 COSM6048207 |
559 | V>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1696954338 | 566 | R>C | No | Ensembl | |
|
rs771164485 COSM1669981 COSM1669979 |
566 | R>H | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1297364570 | 567 | K>Q | No | gnomAD | |
| rs1696953165 | 569 | L>F | No | TOPMed | |
| rs1367195012 | 571 | R>G | No |
TOPMed gnomAD |
|
| rs773319025 | 571 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1367195012 | 571 | R>W | No |
TOPMed gnomAD |
|
| rs2125210809 | 573 | M>V | No | Ensembl | |
| rs138821329 | 575 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1205441472 | 576 | E>K | No | TOPMed | |
| rs1475538215 | 577 | F>L | No | gnomAD | |
| rs2125210628 | 579 | G>C | No | Ensembl | |
| rs996069119 | 580 | N>S | No |
TOPMed gnomAD |
|
| rs996069119 | 580 | N>T | No |
TOPMed gnomAD |
|
| rs141779654 | 581 | R>L | No |
ESP ExAC gnomAD |
|
| rs141779654 | 581 | R>Q | No |
ESP ExAC gnomAD |
|
| rs1559628616 | 583 | K>Q | No | TOPMed | |
| TCGA novel | 584 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779920611 | 585 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs900390606 | 585 | F>V | No |
TOPMed gnomAD |
|
| rs1275244031 | 586 | N>H | No | gnomAD | |
| rs2125210360 | 586 | N>T | No | Ensembl | |
| rs375953005 | 587 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs375953005 | 587 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs764653594 | 587 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs753411185 | 588 | D>E | No |
ExAC TOPMed gnomAD |
|
|
rs1470726208 COSM1733101 |
589 | V>M | pancreas [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
COSM1038148 rs768036999 COSM4868696 |
590 | L>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs759941371 | 591 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs766845024 | 592 | C>W | No |
ExAC TOPMed gnomAD |
|
|
COSM116798 rs763569888 |
593 | V>I | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs763569888 | 593 | V>L | No |
ExAC TOPMed gnomAD |
|
|
COSM3720568 rs1410621879 |
594 | V>M | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1200171137 | 595 | D>N | No | TOPMed | |
| rs994162550 | 596 | E>K | No |
TOPMed gnomAD |
|
| rs137863403 | 597 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs137863403 | 597 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1185419571 | 599 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1696693162 | 600 | S>F | No | TOPMed | |
| rs1461947413 | 600 | S>P | No |
TOPMed gnomAD |
|
| rs1208275232 | 601 | T>N | No | gnomAD | |
| rs1286060196 | 602 | M>T | No | gnomAD | |
| rs1467465738 | 602 | M>V | No | TOPMed | |
| rs1240816829 | 604 | L>V | No | gnomAD | |
| rs555858292 | 607 | A>S | No |
1000Genomes ExAC gnomAD |
|
| rs1301968555 | 613 | A>T | No | gnomAD | |
| rs776067968 | 614 | H>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 614 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs965023055 | 616 | R>C | No |
TOPMed gnomAD |
|
| rs201786310 | 616 | R>H | No |
1000Genomes TOPMed gnomAD |
|
| rs1696688527 | 618 | Q>R | No | Ensembl | |
| rs748747836 | 623 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs144801312 | 626 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1445384593 | 626 | R>W | No |
TOPMed gnomAD |
|
| TCGA novel | 628 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1243666896 | 630 | A>V | No |
TOPMed gnomAD |
|
| TCGA novel | 632 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1696535721 | 633 | Q>H | No | Ensembl | |
| rs752940157 | 634 | R>C | No |
ExAC gnomAD |
|
| rs767579651 | 634 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1210534529 | 638 | C>W | No |
TOPMed gnomAD |
|
| rs1575908815 | 639 | N>S | No | Ensembl | |
|
rs35319679 VAR_051927 |
640 | P>S | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs766183043 | 641 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1696532670 | 642 | V>M | No | gnomAD | |
| rs941280523 | 643 | V>L | No | Ensembl | |
| rs1053747701 | 644 | R>Q | No |
TOPMed gnomAD |
|
| rs1696531925 | 644 | R>W | No | Ensembl | |
|
COSM3695811 rs754235473 |
647 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs769457098 | 647 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1696530492 | 648 | N>K | No | TOPMed | |
| rs766638538 | 649 | P>A | No |
ExAC gnomAD |
|
| rs768263052 | 649 | P>L | No |
ExAC gnomAD |
|
| rs766638538 | 649 | P>S | No |
ExAC gnomAD |
|
|
COSM3587283 COSM3587285 |
651 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746663650 | 654 | I>V | No |
ExAC gnomAD |
|
| rs1696528545 | 656 | A>S | No | Ensembl | |
| TCGA novel | 658 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 659 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1559623072 | 665 | D>N | No | Ensembl | |
| rs1696525614 | 666 | M>I | No | Ensembl | |
| rs1393618300 | 666 | M>R | No | gnomAD | |
| rs1393618300 | 666 | M>T | No | gnomAD | |
| rs1696525170 | 669 | P>T | No | TOPMed | |
| rs1696524669 | 670 | N>D | No |
TOPMed gnomAD |
|
| rs376832198 | 670 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1575908454 | 671 | V>A | No | Ensembl | |
| rs1696523840 | 671 | V>I | No | TOPMed | |
| rs781326232 | 672 | K>R | No | ExAC | |
| rs1696522391 | 673 | P>L | No | Ensembl | |
| rs1696522391 | 673 | P>R | No | Ensembl | |
| rs201571499 | 674 | E>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751783194 | 674 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs762792731 | 675 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs750405546 | 675 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs762792731 | 675 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1452302419 | 678 | K>Q | No | Ensembl | |
| rs1696519267 | 680 | E>D | No |
TOPMed gnomAD |
|
| rs776065570 | 681 | D>G | No |
ExAC gnomAD |
|
| rs76354202 | 683 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs368596372 | 684 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs775169070 | 685 | N>K | No |
ExAC gnomAD |
|
| rs1241838041 | 687 | R>Q | No | gnomAD | |
| rs758753339 | 688 | G>D | No |
ExAC gnomAD |
|
| rs778893195 | 691 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs200821842 | 692 | G>C | No | Ensembl | |
| rs1206174746 | 692 | G>D | No | TOPMed | |
| rs757036944 | 693 | E>D | No |
ExAC gnomAD |
|
| rs1696020351 | 693 | E>G | No | TOPMed | |
|
COSM6010817 COSM6010819 |
693 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753745986 | 694 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs371136421 | 696 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed |
| rs371136421 | 696 | P>T | No |
ESP TOPMed |
|
| rs200895339 | 697 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs200895339 | 697 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs755586675 | 697 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs755586675 | 697 | R>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 697 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1198932533 | 698 | E>A | No |
TOPMed gnomAD |
|
| rs1262891327 | 698 | E>D | No | gnomAD | |
| rs1045230493 | 699 | M>L | No |
TOPMed gnomAD |
|
| rs752273721 | 699 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2125127234 | 701 | M>I | No | Ensembl | |
| rs1481452882 | 702 | G>E | No | TOPMed | |
| rs541280755 | 702 | G>R | No |
1000Genomes ExAC gnomAD |
|
| rs1696015910 | 703 | I>L | No | TOPMed | |
| rs762188644 | 706 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs777017219 | 706 | R>L | No |
ExAC gnomAD |
|
| rs777017219 | 706 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs762188644 | 706 | R>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 707 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs141163963 | 708 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs199955395 | 708 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs199955395 | 708 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs199955395 | 708 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs141163963 | 708 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs746124862 | 709 | K>M | No |
ExAC gnomAD |
|
| rs772248025 | 709 | K>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 710 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1351483270 | 710 | R>G | No |
TOPMed gnomAD |
|
| rs202207235 | 710 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202207235 | 710 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs923910893 | 711 | E>A | No | Ensembl | |
| TCGA novel | 711 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1387907971 | 711 | E>Q | No |
TOPMed gnomAD |
|
| rs1465773678 | 713 | K>N | No | gnomAD | |
| rs756132332 | 713 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs756132332 | 713 | K>T | No |
ExAC TOPMed gnomAD |
|
|
rs145903276 COSM71191 |
715 | N>S | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs947303351 | 716 | E>D | No |
TOPMed gnomAD |
|
| rs1275979123 | 718 | H>Q | No | Ensembl | |
| TCGA novel | 723 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1479297294 | 724 | K>T | No | gnomAD | |
| rs562067887 | 727 | K>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1696008104 | 728 | L>F | No | gnomAD | |
| rs1449978702 | 729 | I>T | No | gnomAD | |
| rs1260051835 | 730 | V>G | No | gnomAD | |
| rs370757999 | 731 | G>A | No |
ESP ExAC gnomAD |
|
| rs1286830134 | 732 | K>E | No |
TOPMed gnomAD |
|
| rs200169490 | 734 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1575888845 | 734 | P>T | No | Ensembl | |
| rs1695955049 | 735 | I>V | No | Ensembl | |
| rs1451937484 | 736 | G>E | No | gnomAD | |
| rs189762650 | 736 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1038145 COSM4866802 |
738 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1348247532 | 738 | L>P | No | TOPMed | |
| rs1695952960 | 739 | H>P | No | TOPMed | |
| rs888566960 | 739 | H>Q | No | TOPMed | |
| rs768073961 | 740 | P>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 741 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1209090686 | 741 | G>W | No | gnomAD | |
| rs1484559344 | 742 | L>F | No |
TOPMed gnomAD |
|
| rs774549995 | 743 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs774549995 | 743 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1575886528 | 744 | C>W | No | Ensembl | |
| rs1575880026 | 745 | V>G | No | Ensembl | |
| rs1425512339 | 745 | V>M | No |
TOPMed gnomAD |
|
| rs751976525 | 746 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1235129260 | 747 | S>C | No | TOPMed | |
| TCGA novel | 748 | L>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2125108238 | 749 | P>L | No | Ensembl | |
| TCGA novel | 751 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs773493024 COSM1418747 |
751 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs909321009 | 752 | R>Q | No | gnomAD | |
|
rs983356831 COSM4113412 |
752 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs769936189 | 753 | L>F | No |
ExAC gnomAD |
|
| rs1359340589 | 753 | L>V | No | gnomAD | |
| rs1186386031 | 753 | L>W | No |
TOPMed gnomAD |
|
| rs1319515707 | 756 | Y>C | No | gnomAD | |
| rs951875351 | 758 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs991492074 | 758 | R>W | No | Ensembl | |
| rs1259140127 | 759 | L>P | No | gnomAD | |
| rs1218464104 | 760 | F>C | No | gnomAD | |
| rs1306096995 | 760 | F>L | No | gnomAD | |
| rs111597884 | 761 | E>G | No | Ensembl | |
| rs762017229 | 762 | V>A | No |
ExAC gnomAD |
|
| rs1695763236 | 764 | D>H | No | TOPMed | |
| rs1695762777 | 765 | P>L | No | TOPMed | |
| rs1387846801 | 765 | P>T | No | gnomAD | |
| rs1367013560 | 768 | P>L | No |
TOPMed gnomAD |
|
| rs1575879576 | 769 | Q>E | No | Ensembl | |
| rs1695762054 | 769 | Q>H | No | Ensembl | |
| rs1423476717 | 772 | G>E | No |
TOPMed gnomAD |
|
| rs776467062 | 772 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs201364726 | 773 | L>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM728511 | 775 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 776 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775243385 | 776 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs771625996 | 776 | R>Q | No |
ExAC gnomAD |
|
| rs745309861 | 777 | E>* | No |
ExAC gnomAD |
|
| rs1366329032 | 777 | E>A | No | gnomAD | |
| rs778484222 | 777 | E>D | No |
ExAC gnomAD |
|
| rs1695758053 | 784 | L>F | No |
TOPMed gnomAD |
|
| rs1238452161 | 786 | V>L | No | gnomAD | |
| rs1575872809 | 787 | V>G | No | Ensembl | |
| TCGA novel | 788 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1695576887 | 790 | I>V | No | TOPMed | |
| rs775502906 | 791 | F>L | No |
ExAC gnomAD |
|
| rs967292167 | 795 | K>N | No |
TOPMed gnomAD |
|
| rs771973106 | 795 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs771973106 | 795 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs745809088 | 796 | N>K | No |
ExAC gnomAD |
|
| rs761936280 | 797 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs777516598 | 799 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs747267384 | 801 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs747267384 | 801 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1332327407 | 803 | R>* | No |
TOPMed gnomAD |
|
| rs994592360 | 803 | R>P | No |
TOPMed gnomAD |
|
| rs994592360 | 803 | R>Q | No |
TOPMed gnomAD |
|
| rs1695571914 | 804 | Q>K | No |
TOPMed gnomAD |
|
| rs758886993 | 805 | S>P | No |
ExAC gnomAD |
|
| rs1157441129 | 806 | F>I | No |
TOPMed gnomAD |
|
| rs750917638 | 806 | F>L | No |
ExAC gnomAD |
|
| rs1157441129 | 806 | F>L | No |
TOPMed gnomAD |
|
| rs1468300503 | 806 | F>S | No | gnomAD | |
| rs376936605 | 810 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1695569465 | 811 | M>T | No | TOPMed | |
| rs1695569686 | 811 | M>V | No | TOPMed | |
| rs1219443751 | 812 | Q>* | No | gnomAD | |
| rs1283683670 | 813 | V>I | No | gnomAD | |
| TCGA novel | 814 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146668270 | 816 | F>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs774833863 | 817 | E>K | No | gnomAD | |
| rs1575872344 | 818 | N>K | No | Ensembl | |
| rs1695567496 | 819 | Q>H | No | Ensembl | |
| rs80261363 | 820 | Y>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3587275 | 821 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772338671 | 822 | P>H | No |
ExAC gnomAD |
|
| rs772338671 | 822 | P>R | No |
ExAC gnomAD |
|
| rs1695347992 | 822 | P>S | No |
TOPMed gnomAD |
|
| rs1247397549 | 823 | N>H | No |
TOPMed gnomAD |
|
| rs764427757 | 823 | N>K | No |
TOPMed gnomAD |
|
| rs149003329 | 823 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779245783 | 825 | I>T | No |
ExAC gnomAD |
|
| rs149771840 | 826 | R>Q | No |
ESP ExAC gnomAD |
|
|
rs1695345942 COSM295085 |
826 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs1695344551 | 828 | T>I | No | TOPMed | |
| rs553936985 | 829 | S>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3587273 | 830 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756181588 | 832 | P>S | No |
ExAC gnomAD |
|
| rs139635176 | 833 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs754821070 | 835 | D>G | No |
ExAC gnomAD |
|
| rs1695342923 | 835 | D>N | No | Ensembl | |
| rs751462955 | 836 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs751462955 | 836 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs201050159 | 837 | K>E | No | Ensembl | |
| rs766262541 | 838 | V>A | No |
ExAC gnomAD |
|
| rs1695341509 | 838 | V>L | No | TOPMed | |
|
COSM4838857 COSM4838859 |
839 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs750352579 | 840 | I>L | No |
ExAC gnomAD |
|
| rs1051408367 | 843 | N>D | No | Ensembl | |
| rs1051408367 | 843 | N>H | No | Ensembl | |
| rs1319696371 | 845 | P>A | No |
TOPMed gnomAD |
|
| rs776155190 | 846 | N>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 846 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763203754 | 850 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs371733963 | 850 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs369092436 | 851 | K>Q | No |
ESP TOPMed gnomAD |
|
| rs1695337342 | 852 | K>I | No | gnomAD | |
|
COSM1038137 rs2125074583 |
853 | F>L | endometrium Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated Ensembl NCI-TCGA Cosmic |
| rs1173528131 | 854 | T>I | No |
TOPMed gnomAD |
|
| rs749827851 | 855 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs749827851 | 855 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1179510311 | 856 | D>H | No | gnomAD | |
| rs769774476 | 858 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1232118149 | 858 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM3060451 | 860 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1281037330 | 861 | I>T | No | gnomAD | |
| rs181893318 | 861 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781123855 | 862 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs1205326935 | 862 | A>P | No |
TOPMed gnomAD |
|
| rs781123855 | 862 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs747210938 COSM1308553 |
863 | E>K | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1224024694 | 866 | E>Q | No | gnomAD | |
| rs901017895 | 867 | M>I | No | TOPMed | |
| rs1695332074 | 867 | M>K | No | Ensembl | |
| rs1040883802 | 868 | E>G | No | TOPMed | |
| rs780035266 | 870 | H>Y | No |
ExAC gnomAD |
|
| rs1695331006 | 872 | I>V | No |
TOPMed gnomAD |
|
|
COSM6163235 COSM6163237 |
873 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1451166177 | 873 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1451166177 | 873 | E>Q | No |
TOPMed gnomAD |
|
|
COSM1254914 rs776819752 |
874 | S>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs748587727 | 874 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1343496963 | 875 | E>G | No | gnomAD | |
| rs747265961 | 877 | E>G | No |
ExAC gnomAD |
|
| rs1462102870 | 877 | E>K | No | gnomAD | |
| COSM6163238 | 877 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1038136 | 879 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753690784 | 883 | V>A | No |
ExAC gnomAD |
|
| rs562700735 | 883 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs562700735 | 883 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4783484 COSM1210866 |
884 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1559607247 | 884 | R>Q | No | Ensembl | |
| rs201800793 | 884 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs372297620 | 885 | P>L | No |
ESP ExAC gnomAD |
|
| rs1340464304 | 885 | P>S | No |
TOPMed gnomAD |
|
| rs1695227273 | 886 | S>N | No | TOPMed | |
| rs1695226639 | 887 | M>I | No | Ensembl | |
| rs1272965401 | 887 | M>V | No |
TOPMed gnomAD |
|
| rs368203899 | 888 | S>C | No |
ESP TOPMed gnomAD |
|
| COSM269384 | 889 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1575860491 | 890 | C>G | No | Ensembl | |
| rs1695225692 | 890 | C>Y | No | Ensembl | |
| rs1575860478 | 891 | S>F | No | Ensembl | |
| rs145769651 | 892 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1446419360 | 892 | S>R | No |
TOPMed gnomAD |
|
| rs145769651 | 892 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1695224042 | 894 | K>R | No | gnomAD | |
| rs750854478 | 895 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs765493111 | 896 | E>K | No |
ExAC gnomAD |
|
| rs765493111 | 896 | E>Q | No |
ExAC gnomAD |
|
| rs776962116 | 897 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs776962116 | 897 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs1163017684 | 898 | G>S | No | gnomAD | |
| rs772481223 | 900 | G>R | No |
ExAC gnomAD |
|
| rs747094094 | 902 | L>P | No |
ExAC gnomAD |
|
| rs1695220498 | 903 | S>R | No | TOPMed | |
| rs138048919 | 904 | R>Q | No |
ESP TOPMed |
|
| rs374319490 | 904 | R>W | No |
ESP ExAC gnomAD |
|
| rs749272326 | 905 | A>V | No |
ExAC gnomAD |
|
| rs1468518803 | 906 | C>Y | No |
TOPMed gnomAD |
|
| rs1327484270 | 907 | L>P | No | TOPMed | |
| rs1327484270 | 907 | L>Q | No | TOPMed | |
| TCGA novel | 909 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755629839 | 909 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1695217850 | 910 | S>R | No | gnomAD | |
| rs2125067001 | 910 | S>T | No | Ensembl | |
| rs752267527 | 911 | Y>C | No |
ExAC gnomAD |
|
| rs1695217049 | 912 | A>V | No | Ensembl | |
| rs751139467 | 914 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs751139467 | 914 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs765664329 | 916 | G>A | No | ExAC | |
| rs1575860101 | 916 | G>S | No | Ensembl | |
| rs1370330591 | 919 | R>C | No |
TOPMed gnomAD |
|
| rs1324202560 | 919 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1348590310 | 920 | S>N | No | gnomAD | |
| rs754408494 | 921 | A>S | No |
ExAC gnomAD |
|
|
COSM4624957 COSM4624959 rs754408494 |
921 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1695214387 | 921 | A>V | No | TOPMed | |
| rs1286286662 | 922 | L>V | No |
TOPMed gnomAD |
|
| rs2125066760 | 923 | S>G | No | Ensembl | |
| rs1695213659 | 923 | S>N | No | Ensembl | |
| rs1695213407 | 924 | S>G | No | Ensembl | |
| rs560424556 | 927 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201498408 | 927 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs560424556 | 927 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs761827844 COSM4113411 COSM4113409 |
930 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1695211176 | 931 | E>G | No | TOPMed | |
| rs1420465801 | 932 | A>G | No | gnomAD | |
| rs1321664446 | 932 | A>T | No |
TOPMed gnomAD |
|
| rs1420465801 | 932 | A>V | No | gnomAD | |
| rs1195637536 | 933 | G>R | No |
TOPMed gnomAD |
|
| COSM6096276 | 933 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1294863415 COSM3060428 |
935 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1415351354 | 935 | R>Q | No | gnomAD | |
|
rs1694511803 RCV001264653 |
936 | G>R | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1694511803 | 936 | G>W | No |
TOPMed gnomAD |
|
|
rs770811609 COSM1038135 |
937 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1354797446 | 937 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1354797446 | 937 | R>P | No |
TOPMed gnomAD |
|
| rs1185580258 | 938 | R>C | No | TOPMed | |
| rs994980836 | 938 | R>H | No |
TOPMed gnomAD |
|
| rs1694510260 | 939 | S>N | No | Ensembl | |
| rs139603221 | 941 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs758268841 | 943 | S>* | No |
TOPMed gnomAD |
|
| rs758268841 | 943 | S>L | No |
TOPMed gnomAD |
|
| rs1166043335 | 945 | E>D | No |
TOPMed gnomAD |
|
| rs1694507612 | 945 | E>Q | No | Ensembl | |
| rs1475070515 | 946 | S>T | No |
TOPMed gnomAD |
|
| rs779603962 | 947 | N>D | No |
ExAC gnomAD |
|
| rs779603962 | 947 | N>H | No |
ExAC gnomAD |
|
| rs758035076 | 947 | N>K | No |
ExAC gnomAD |
|
| rs1447672339 | 948 | V>M | No | gnomAD | |
| rs745487079 | 949 | E>K | No |
ExAC gnomAD |
|
| rs2124954778 | 950 | F>L | No | Ensembl | |
| rs1693994045 | 951 | Q>L | No | TOPMed | |
| rs753402468 | 952 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1174425174 | 952 | P>T | No |
TOPMed gnomAD |
|
| rs1163980959 | 953 | F>C | No | gnomAD | |
| rs755799875 | 953 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1350469946 | 953 | F>V | No | gnomAD | |
| rs568360461 | 954 | E>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1188835435 | 954 | E>D | No | gnomAD | |
| rs568360461 | 954 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs568360461 | 954 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs766792546 | 955 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs765914797 | 956 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs200584672 | 956 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1265429397 | 957 | Q>* | No | gnomAD | |
| rs149929550 | 958 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs149929550 | 958 | P>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1275382109 | 959 | S>L | No | gnomAD | |
| rs779327139 | 960 | V>L | No |
TOPMed gnomAD |
|
| rs779327139 | 960 | V>M | No |
TOPMed gnomAD |
|
| rs761008969 | 961 | L>P | No |
ExAC gnomAD |
|
| rs1693988987 | 962 | C>Y | No | Ensembl | |
| rs2124954159 | 963 | S>C | No | Ensembl | |
| rs775517449 | 963 | S>T | No |
ExAC gnomAD |
|
| rs745714782 | 964 | S>K | No |
ExAC gnomAD |
No associated diseases with Q6DN90
8 regional properties for Q6DN90
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | HEAT, type 2 | 169 - 207 | IPR021133-1 |
| repeat | HEAT, type 2 | 1414 - 1452 | IPR021133-2 |
| domain | CLASP N-terminal domain | 316 - 536 | IPR024395 |
| domain | TOG domain | 2 - 234 | IPR034085-1 |
| domain | TOG domain | 312 - 548 | IPR034085-2 |
| domain | TOG domain | 836 - 1072 | IPR034085-3 |
| domain | TOG domain | 1251 - 1477 | IPR034085-4 |
| domain | XMAP215/Dis1/CLASP, TOG domain | 66 - 204 | IPR048491 |
Functions
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| guanyl-nucleotide exchange factor activity | Stimulates the exchange of GDP to GTP on a signaling GTPase, changing its conformation to its active form. Guanine nucleotide exchange factors (GEFs) act by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP), which is more abundant in the cell under normal cellular physiological conditions. |
| lipid binding | Binding to a lipid. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| dendritic spine development | The process whose specific outcome is the progression of the dendritic spine over time, from its formation to the mature structure. A dendritic spine is a protrusion from a dendrite and a specialized subcellular compartment involved in synaptic transmission. |
| positive regulation of focal adhesion disassembly | Any process that activates or increases the frequency, rate or extent of disaggregation of a focal adhesion into its constituent components. |
| positive regulation of keratinocyte migration | Any process that activates or increases the frequency, rate or extent of keratinocyte migration. |
| regulation of ARF protein signal transduction | Any process that modulates the frequency, rate or extent of ARF protein signal transduction. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9Y6D5 | ARFGEF2 | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | Homo sapiens (Human) | PR |
| Q9Y6D6 | ARFGEF1 | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 | Homo sapiens (Human) | PR |
| Q5JU85 | IQSEC2 | IQ motif and SEC7 domain-containing protein 2 | Homo sapiens (Human) | EV |
| Q9UPP2 | IQSEC3 | IQ motif and SEC7 domain-containing protein 3 | Homo sapiens (Human) | SS |
| Q99418 | CYTH2 | Cytohesin-2 | Homo sapiens (Human) | SS |
| Q9UIA0 | CYTH4 | Cytohesin-4 | Homo sapiens (Human) | SS |
| O43739 | CYTH3 | Cytohesin-3 | Homo sapiens (Human) | EV |
| Q15438 | CYTH1 | Cytohesin-1 | Homo sapiens (Human) | SS |
| Q92538 | GBF1 | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 | Homo sapiens (Human) | PR |
| Q5DU25 | Iqsec2 | IQ motif and SEC7 domain-containing protein 2 | Mus musculus (Mouse) | SS |
| Q3TES0 | Iqsec3 | IQ motif and SEC7 domain-containing protein 3 | Mus musculus (Mouse) | SS |
| Q8R0S2 | Iqsec1 | IQ motif and SEC7 domain-containing protein 1 | Mus musculus (Mouse) | SS |
| Q76M68 | Iqsec3 | IQ motif and SEC7 domain-containing protein 3 | Rattus norvegicus (Rat) | SS |
| A0A0G2JUG7 | Iqsec1 | IQ motif and SEC7 domain-containing protein 1 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MWCLHCNSER | TQSLLELELD | SGVEGEAPSS | ETGTSLDSPS | AYPQGPLVPG | SSLSPDHYEH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TSVGAYGLYS | GPPGQQQRTR | RPKLQHSTSI | LRKQAEEEAI | KRSRSLSESY | ELSSDLQDKQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VEMLERKYGG | RLVTRHAART | IQTAFRQYQM | NKNFERLRSS | MSENRMSRRI | VLSNMRMQFS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FEGPEKVHSS | YFEGKQVSVT | NDGSQLGALV | SPECGDLSEP | TTLKSPAPSS | DFADAITELE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DAFSRQVKSL | AESIDDALNC | RSLHTEEAPA | LDAARARDTE | PQTALHGMDH | RKLDEMTASY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SDVTLYIDEE | ELSPPLPLSQ | AGDRPSSTES | DLRLRAGGAA | PDYWALAHKE | DKADTDTSCR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| STPSLERQEQ | RLRVEHLPLL | TIEPPSDSSV | DLSDRSERGS | LKRQSAYERS | LGGQQGSPKH |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GPHSGAPKSL | PREEPELRPR | PPRPLDSHLA | INGSANRQSK | SESDYSDGDN | DSINSTSNSN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DTINCSSESS | SRDSLREQTL | SKQTYHKEAR | NSWDSPAFSN | DVIRKRHYRI | GLNLFNKKPE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| KGVQYLIERG | FVPDTPVGVA | HFLLQRKGLS | RQMIGEFLGN | RQKQFNRDVL | DCVVDEMDFS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| TMELDEALRK | FQAHIRVQGE | AQKVERLIEA | FSQRYCICNP | GVVRQFRNPD | TIFILAFAII |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LLNTDMYSPN | VKPERKMKLE | DFIKNLRGVD | DGEDIPREML | MGIYERIRKR | ELKTNEDHVS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| QVQKVEKLIV | GKKPIGSLHP | GLGCVLSLPH | RRLVCYCRLF | EVPDPNKPQK | LGLHQREIFL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| FNDLLVVTKI | FQKKKNSVTY | SFRQSFSLYG | MQVLLFENQY | YPNGIRLTSS | VPGADIKVLI |
| 850 | 860 | 870 | 880 | 890 | 900 |
| NFNAPNPQDR | KKFTDDLRES | IAEVQEMEKH | RIESELEKQK | GVVRPSMSQC | SSLKKESGNG |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TLSRACLDDS | YASGEGLKRS | ALSSSLRDLS | EAGKRGRRSS | AGSLESNVEF | QPFEPLQPSV |
| LCS |