Q6DE55
Gene name |
arhgap45 |
Protein name |
Rho GTPase-activating protein 45 |
Names |
Minor histocompatibility protein HA-1 |
Species |
Xenopus laevis (African clawed frog) |
KEGG Pathway |
xla:446507 |
EC number |
|
Protein Class |
|
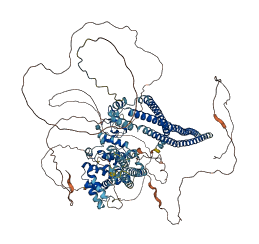
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
670-1107 (C1-GAP-tail domains) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q6DE55
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6DE55-F1 | Predicted | AlphaFoldDB |
No variants for Q6DE55
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q6DE55 | |||||
No associated diseases with Q6DE55
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTPase activator activity | Binds to and increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP. |
| metal ion binding | Binding to a metal ion. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MFSRKKRELM | KTPSLSKKSR | AGSPAPQNDL | TRKDVTDSSN | DLASSPPSNS | SPVSSGTLKR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PSSLSRHASA | AGIPLSSPRG | KATKPASTPS | PPESGEGPFI | DVEDISQLLG | DVARFAERLE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KLRDVVQDEE | LKETRRPLAH | ECLGEALRLL | RQVINKYPLL | NTVETLTAAG | TLISKVKGFH |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YESSIENDKR | DFEKALESMA | VCFSSTISEF | LMGEVDSSTL | LSLPPGDQSQ | SMESLCGGLS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GGEGALPSAH | EYVEAGGHLG | EDVDVILQRS | DGGVQAALLY | AKNMAKYLKD | LSSYIEKRTI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LEMEYAKGLQ | KLVNAYKGTL | NQETHMPFQS | IYSVALEQDL | EHGHGILHTA | LTLQHQTFLQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PINMRRQEHE | KRRKEVKEQW | QRAQRKLMEA | ESNLRKARQA | YMQRSEEHER | ALYNATRAEE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| EQSHSGTRSL | DKKRRAEEEA | KNRAEEAMAT | YRTCIADAKT | QKQELEDVKV | NVLRQLQELI |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KQSDQILRSA | TISYYQSMHM | QTAPLPVGFQ | MLCESSKLYD | LGQQYASYVR | QLGAVNEPET |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SYDFQPYTPQ | ITWSPCIRAR | KSSFNSQDIP | SSENKEISGE | ERGVERRGGR | GHQVHKSWPT |
| 610 | 620 | 630 | 640 | 650 | 660 |
| AITEGDPAVS | SATVPAFPEK | LHQPLSPTEN | VDQKRLSASF | EQSINGLSGS | LEVQNSTGPF |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RNIGLSRAAL | THRLRKLRTP | SKCRECNSYV | YFQGAECEEC | SLACHKKCLE | TLAIQCGHKK |
| 730 | 740 | 750 | 760 | 770 | 780 |
| LQGRLLLFGR | DFSETALRSP | DHIPFLIRKC | VSEIEERALI | MKGIYRVNGV | KTRVEKLCQA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| FENGKELVEL | SQASPHDLSN | VLKLYLRQLP | EPLIPFRLYN | GLMGLAKESL | RGTETGKGPR |
| 850 | 860 | 870 | 880 | 890 | 900 |
| LQDKGPNTET | DVLSIVVQLK | ELLQDLPSEN | RTTLQYLVKH | LCRVSEQEQL | NKMSPSNLGI |
| 910 | 920 | 930 | 940 | 950 | 960 |
| VFGPALMRPR | PTDATVSLSS | LVDYPHQARI | VETLIIFYST | IFQEPVSNTD | IGTGNSSSDD |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| TASMQSRARL | QVTVEEDLSE | LTPEYQIPVF | KEPGASTVES | DSESDGAEDI | PGTWKPQTTR |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| GHLTKEASVT | SAEDIPYIEG | EAQSESEEDR | DQTQENLAEN | NTNQSNNVAV | NGHCCVPHFH |
| 1090 | 1100 | ||||
| CHTQLPAIRM | MHGKIYVSSA | DRRPHFV |