Q6DCD7
Gene name |
zcchc4 |
Protein name |
rRNA N6-adenosine-methyltransferase ZCCHC4 |
Names |
Zinc finger CCHC domain-containing protein 4 |
Species |
Xenopus laevis (African clawed frog) |
KEGG Pathway |
xla:447056 |
EC number |
|
Protein Class |
|
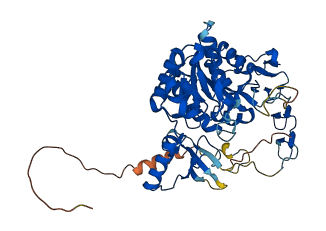
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q6DCD7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q6DCD7-F1 | Predicted | AlphaFoldDB |
No variants for Q6DCD7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q6DCD7 | |||||
No associated diseases with Q6DCD7
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| nucleic acid binding | Binding to a nucleic acid. |
| rRNA (adenine-N6-)-methyltransferase activity | Catalysis of the reaction: S-adenosyl-L-methionine + rRNA = S-adenosyl-L-homocysteine + rRNA containing N6-methyladenine. |
| S-adenosyl-L-methionine binding | Binding to S-adenosyl-L-methionine. |
| zinc ion binding | Binding to a zinc ion (Zn). |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| positive regulation of translation | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| rRNA methylation | The posttranscriptional addition of methyl groups to specific residues in an rRNA molecule. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEAAEGSENY | DGIQVLLSRE | VIDTAPQCPH | GPTLLFVKVS | QGKEQGRRFY | ACSACRDRKD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CHFFQWEDEK | VSQARLAARE | EYNKSHQPPM | THAEYARSFQ | EFTALPLAKR | KFCCDCQQLL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LQTNWETHSR | HRVLGDISLS | QLKRPSQLLH | PLENKKANAQ | YLFADRSCTF | LLDTIIALGF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RRVLCVGTPR | LHELIKLRAC | KGTTPPIKSL | LLDIDFRYSQ | FYSEEEFSHY | NMFNHHFFGG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EAAKLVCQKF | LQEEDGNGAL | LVTDPPFGGL | VEPLAFSFKR | LREMWKDSNP | ENANNLPIFW |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MFPYFFESRI | LQCFPDFAML | DYQVDYDNHA | LYKHGKTGRK | QSPVRIFTDL | PLDKIVLPAS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EGYRFCSVCE | RFVCSENKHC | DICNRCTSKD | GRSWKHCSQC | KKCVKPSWSH | CSACNHCALP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GHPCGTAGDG | CFLCGGKGHK | RRGCPHLSVS | ADGERMKRNL | KQRSIKGNMK | KQPTATTSKK |
| KKRKRNNPC |