Q66X01
Gene name |
Nlrp9c (Nalp9c) |
Protein name |
NACHT, LRR and PYD domains-containing protein 9C |
Names |
NALP-zeta |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:330490 |
EC number |
|
Protein Class |
|
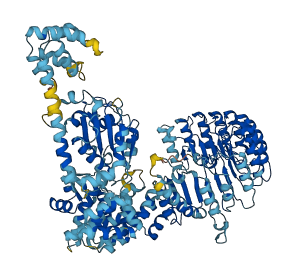
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q66X01
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q66X01-F1 | Predicted | AlphaFoldDB |
88 variants for Q66X01
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388870469 | 27 | L>H | No | EVA | |
| rs245274055 | 32 | Q>P | No | EVA | |
| rs258573300 | 55 | T>M | No | EVA | |
| rs3388870407 | 75 | Q>H | No | EVA | |
| rs3388883160 | 109 | T>S | No | EVA | |
| rs3388892579 | 113 | N>D | No | EVA | |
| rs16790590 | 123 | T>I | No | EVA | |
| rs16790590 | 123 | T>N | No | EVA | |
| rs3388888505 | 159 | L>* | No | EVA | |
| rs234972737 | 165 | D>N | No | EVA | |
| rs261219695 | 177 | Q>R | No | EVA | |
| rs3388888596 | 181 | F>V | No | EVA | |
| rs3388883119 | 182 | F>I | No | EVA | |
| rs3388888698 | 195 | L>S | No | EVA | |
| rs3388873509 | 197 | E>K | No | EVA | |
| rs3388883172 | 210 | L>M | No | EVA | |
| rs16790584 | 218 | K>T | No | EVA | |
| rs222344053 | 222 | F>I | No | EVA | |
| rs3397679226 | 252 | N>I | No | EVA | |
| rs3397658262 | 253 | V>I | No | EVA | |
| rs3397742102 | 256 | S>N | No | EVA | |
| rs3397689260 | 256 | S>R | No | EVA | |
| rs3388849741 | 263 | L>F | No | EVA | |
| rs3388864579 | 271 | E>D | No | EVA | |
| rs3388888780 | 276 | S>I | No | EVA | |
| rs16790585 | 278 | S>P | No | EVA | |
| rs3388888508 | 298 | Q>L | No | EVA | |
| rs3388895976 | 305 | V>E | No | EVA | |
| rs16790586 | 324 | E>G | No | EVA | |
| rs16790587 | 334 | S>T | No | EVA | |
| rs3388893495 | 347 | H>D | No | EVA | |
| rs16790588 | 347 | H>Q | No | EVA | |
| rs3388882984 | 358 | E>K | No | EVA | |
| rs3388878867 | 371 | A>S | No | EVA | |
| rs3388890832 | 380 | S>L | No | EVA | |
| rs3388883139 | 404 | Y>C | No | EVA | |
| rs3388870428 | 410 | D>N | No | EVA | |
| rs3388888706 | 418 | E>D | No | EVA | |
| rs3388890864 | 487 | A>T | No | EVA | |
| rs3388878932 | 491 | F>L | No | EVA | |
| rs16790580 | 501 | T>A | No | EVA | |
| rs16790581 | 515 | I>V | No | EVA | |
| rs3388881267 | 517 | Q>H | No | EVA | |
| rs3388891371 | 525 | S>G | No | EVA | |
| rs3388888531 | 533 | E>D | No | EVA | |
| rs3388888537 | 541 | L>F | No | EVA | |
| rs16790582 | 542 | F>L | No | EVA | |
| rs16790583 | 546 | I>V | No | EVA | |
| rs3388888774 | 587 | L>S | No | EVA | |
| rs234980939 | 589 | N>T | No | EVA | |
| rs37282684 | 621 | A>V | No | EVA | |
| rs3388888781 | 646 | K>R | No | EVA | |
| rs3388893438 | 653 | C>S | No | EVA | |
| rs221830072 | 661 | N>T | No | EVA | |
| rs3388892596 | 690 | L>R | No | EVA | |
| rs16790595 | 723 | R>G | No | EVA | |
| rs38573551 | 742 | M>I | No | EVA | |
| rs3388849780 | 746 | L>F | No | EVA | |
| rs37239843 | 747 | I>V | No | EVA | |
| rs3397708282 | 754 | L>H | No | EVA | |
| rs3397851501 | 756 | L>F | No | EVA | |
| rs3397708226 | 761 | L>F | No | EVA | |
| rs3397296172 | 764 | K>Q | No | EVA | |
| rs3396914780 | 765 | G>* | No | EVA | |
| rs3388849787 | 771 | K>E | No | EVA | |
| rs217797579 | 797 | H>Y | No | EVA | |
| rs3388883108 | 816 | N>I | No | EVA | |
| rs254461538 | 833 | P>S | No | EVA | |
| rs16790682 | 857 | A>T | No | EVA | |
| rs3397618743 | 881 | K>T | No | EVA | |
| rs16790669 | 882 | R>W | No | EVA | |
| rs3396914770 | 884 | C>* | No | EVA | |
| rs3388883173 | 884 | C>Y | No | EVA | |
| rs3388886251 | 888 | C>S | No | EVA | |
| rs3397742009 | 890 | P>H | No | EVA | |
| rs3397851498 | 891 | K>N | No | EVA | |
| rs3396914806 | 892 | C>R | No | EVA | |
| rs3397679231 | 893 | K>* | No | EVA | |
| rs16790671 | 894 | V>L | No | EVA | |
| rs3388890838 | 904 | L>H | No | EVA | |
| rs38159347 | 906 | N>S | No | EVA | |
| rs3388895932 | 915 | A>S | No | EVA | |
| rs16790660 | 921 | T>P | No | EVA | |
| rs3388890828 | 925 | L>F | No | EVA | |
| rs3388864528 | 938 | V>A | No | EVA | |
| rs3388878870 | 973 | E>K | No | EVA | |
| rs3388891298 | 981 | V>G | No | EVA | |
| rs38180668 | 992 | K>E | No | EVA |
No associated diseases with Q66X01
8 regional properties for Q66X01
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 186 - 444 | IPR000719 |
| domain | EF-hand domain | 487 - 558 | IPR002048-1 |
| domain | EF-hand domain | 559 - 628 | IPR002048-2 |
| active_site | Serine/threonine-protein kinase, active site | 306 - 318 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 192 - 215 | IPR017441 |
| binding_site | EF-Hand 1, calcium-binding site | 500 - 512 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 536 - 548 | IPR018247-2 |
| binding_site | EF-Hand 1, calcium-binding site | 606 - 618 | IPR018247-3 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| inflammasome complex | A cytosolic protein complex that is capable of activating caspase-1. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| blastocyst development | The process whose specific outcome is the progression of the blastocyst over time, from its formation to the mature structure. The mammalian blastocyst is a hollow ball of cells containing two cell types, the inner cell mass and the trophectoderm. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| regulation of inflammatory response | Any process that modulates the frequency, rate or extent of the inflammatory response, the immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. |
3 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8CCN1 | Nlrp10 | NACHT, LRR and PYD domains-containing protein 10 | Mus musculus (Mouse) | PR |
| Q8R4B8 | Nlrp3 | NACHT, LRR and PYD domains-containing protein 3 | Mus musculus (Mouse) | SS |
| E9Q5R7 | Nlrp12 | NACHT, LRR and PYD domains-containing protein 12 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVDSSSYGLL | QYFQKLSDEE | FQRFKELLQK | EQEKFKLKPL | SWTKIKNTSK | EDLVTQLYTH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YPRQVWDMVL | NLFLQVNRKD | LSTMAQIERR | DKQNKYKEFM | KNLFQYIWTS | ETNTYMPDRS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YNTIIDRQYK | ALLDIFDSES | DPATAVVLGT | RGKGKTVFLR | KAMLDWASGV | LLQNRFQYVF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FFSVFSLNNT | TELSLAELIS | SKLPECSETL | DDILSNPKRI | LFVLDGFDYL | KFDLELRTNL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CNDWRKRLPT | QNVLSSLLQK | IMLPECSLLL | ELGESSCSKI | IPLLQNPREI | IMSGLSEQSI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YFYCVSFFKI | QLGVEVFKDL | KKNEPLFTLC | SNPSMLWMIC | SSLMWGHYSR | EEVISSSEST |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SAIHTIFIMS | AFKSIFGLGS | SKYKRFKLKT | LCTLAVEGMW | KQVYVFDSED | LRRNKISESD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KTVWLKMKFL | QIQGNNIMFY | HSTLQWYFAT | LFYFLKQDKD | TYHPVIGSLP | QLLGEIYAHK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QNQWTHAQTF | FFGIATKHVI | TLLKPCFGNI | SFKTIRQEII | RYLKSLSQPE | CNEKLVHPKK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LFFCLIENQE | ERFVSQVMNL | FEEITVDISD | SDDLGAAEYS | LLRASKLKNL | HLHIQKKVFS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EIHDPEYGSL | ENFKLDQKFS | AINWTMLSIL | FCNLHVLDLG | SCHFNKKVIE | VLCNSLSPTP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| NMPLTVFKLQ | RLLCSFMTNF | GDGSLFCTFL | QIPQLKYLNL | YGTDLSNDVV | EMLCSALKCS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| TCRVEELLLG | KCDISSEACG | IMATFLINSK | VKHLSLVENP | LKNKGVMFLC | KMLKDPSCVL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| ESLMLSYCCL | TFIACGHLYE | ALLSNKHLSL | LDLGSNFLED | IGVNLLCEAL | KYPNCTLKEL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| WLPGCYLTSE | CCEEISAVLT | CNKNLKTLKL | GNNNIQDTGV | KRLCEALCHP | KCKVQCLGLD |
| 910 | 920 | 930 | 940 | 950 | 960 |
| MCELSNDCCE | DLALALITCN | TLKSLNLDWN | ALHHSGLVML | CEALNHKKCK | LNMLGLDKSS |
| 970 | 980 | 990 | 1000 | ||
| FSEESQTFLQ | AVEKKNNNLN | VLHFPWVEDE | LKKRGVRLVW | NSKN |