Q66L42
Gene name |
Map3k10 |
Protein name |
Mitogen-activated protein kinase kinase kinase 10 |
Names |
EC 2.7.11.25 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:269881 |
EC number |
2.7.11.25: Protein-serine/threonine kinases |
Protein Class |
|
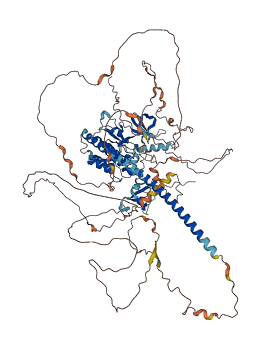
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
444-467 (Proline-rich region) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
247-268 (Activation loop)
Target domain |
98-360 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Du Y et al. (2005) "Cdc42 induces activation loop phosphorylation and membrane targeting of mixed lineage kinase 3", The Journal of biological chemistry, 280, 42984-93
- Zhang H et al. (2001) "Autoinhibition of mixed lineage kinase 3 through its Src homology 3 domain", The Journal of biological chemistry, 276, 45598-603
- Handley ME et al. (2007) "Mixed lineage kinases (MLKs): a role in dendritic cells, inflammation and immunity", International journal of experimental pathology, 88, 111-26
- Kokoszka ME et al. (2018) "Identification of two distinct peptide-binding pockets in the SH3 domain of human mixed-lineage kinase 3", The Journal of biological chemistry, 293, 13553-13565
Autoinhibited structure

Activated structure

1 structures for Q66L42
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q66L42-F1 | Predicted | AlphaFoldDB |
16 variants for Q66L42
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388880902 | 168 | P>T | No | EVA | |
| rs3397762813 | 203 | V>A | No | EVA | |
| rs3388893583 | 277 | L>M | No | EVA | |
| rs3388865861 | 288 | S>C | No | EVA | |
| rs3397667155 | 292 | L>P | No | EVA | |
| rs3397450711 | 298 | T>A | No | EVA | |
| rs3388875021 | 299 | G>R | No | EVA | |
| rs3388893806 | 300 | E>K | No | EVA | |
| rs3388875052 | 370 | L>V | No | EVA | |
| rs3388884050 | 379 | D>G | No | EVA | |
| rs3388892829 | 421 | R>H | No | EVA | |
| rs3388893537 | 463 | R>C | No | EVA | |
| rs3388891585 | 640 | C>Y | No | EVA | |
| rs31866481 | 673 | L>M | No | EVA | |
| rs3388872309 | 718 | R>C | No | EVA | |
| rs3397699160 | 767 | S>T | No | EVA |
No associated diseases with Q66L42
6 regional properties for Q66L42
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 98 - 360 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 98 - 357 | IPR001245 |
| domain | SH3 domain | 16 - 81 | IPR001452 |
| active_site | Serine/threonine-protein kinase, active site | 218 - 230 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 104 - 125 | IPR017441 |
| domain | MLK1-3, SH3 domain | 20 - 76 | IPR035779 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.25 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| JUN kinase kinase kinase activity | Catalysis of the reaction |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction |
| protein serine kinase activity | Catalysis of the reactions |
| transcription corepressor activity | A transcription coregulator activity that represses or decreases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Corepressors often act by altering chromatin structure and modifications. For example, one class of transcription corepressors modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
41 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VDU3 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Bos taurus (Bovine) | SS |
| Q4TVR5 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Bos taurus (Bovine) | PR |
| Q3SZJ2 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Bos taurus (Bovine) | PR |
| Q6XUX0 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Gallus gallus (Chicken) | PR |
| P83104 | Takl1 | Putative mitogen-activated protein kinase kinase kinase 7-like | Drosophila melanogaster (Fruit fly) | PR |
| Q95UN8 | slpr | Mitogen-activated protein kinase kinase kinase | Drosophila melanogaster (Fruit fly) | EV |
| O43353 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Homo sapiens (Human) | PR |
| Q8NB16 | MLKL | Mixed lineage kinase domain-like protein | Homo sapiens (Human) | EV |
| Q16584 | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | Homo sapiens (Human) | EV |
| P00540 | MOS | Proto-oncogene serine/threonine-protein kinase mos | Homo sapiens (Human) | PR |
| Q5TCX8 | MAP3K21 | Mitogen-activated protein kinase kinase kinase 21 | Homo sapiens (Human) | PR |
| Q6XUX3 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Homo sapiens (Human) | PR |
| O43318 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Homo sapiens (Human) | SS |
| Q9NYL2 | MAP3K20 | Mitogen-activated protein kinase kinase kinase 20 | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| P80192 | MAP3K9 | Mitogen-activated protein kinase kinase kinase 9 | Homo sapiens (Human) | SS |
| Q02779 | MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 | Homo sapiens (Human) | SS |
| Q62073 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Mus musculus (Mouse) | EV |
| P00536 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Mus musculus (Mouse) | PR |
| P58801 | Ripk2 | Receptor-interacting serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q3U1V8 | Map3k9 | Mitogen-activated protein kinase kinase kinase 9 | Mus musculus (Mouse) | SS |
| Q80XI6 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Mus musculus (Mouse) | PR |
| Q8VDG6 | Map3k21 | Mitogen-activated protein kinase kinase kinase 21 | Mus musculus (Mouse) | PR |
| P04627 | Araf | Serine/threonine-protein kinase A-Raf | Mus musculus (Mouse) | PR |
| O55222 | Ilk | Integrin-linked protein kinase | Mus musculus (Mouse) | PR |
| Q9ESL4 | Map3k20 | Mitogen-activated protein kinase kinase kinase 20 | Mus musculus (Mouse) | PR |
| Q99N57 | Raf1 | RAF proto-oncogene serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| P28028 | Braf | Serine/threonine-protein kinase B-raf | Mus musculus (Mouse) | SS |
| Q9D2Y4 | Mlkl | Mixed lineage kinase domain-like protein | Mus musculus (Mouse) | SS |
| P0C8E4 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Rattus norvegicus (Rat) | SS |
| Q66HA1 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Rattus norvegicus (Rat) | PR |
| P00539 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Rattus norvegicus (Rat) | PR |
| D3ZG83 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Rattus norvegicus (Rat) | SS |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9FPR3 | EDR1 | Serine/threonine-protein kinase EDR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2MHE4 | HT1 | Serine/threonine/tyrosine-protein kinase HT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q05609 | CTR1 | Serine/threonine-protein kinase CTR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q67E00 | dstyk | Dual serine/threonine and tyrosine protein kinase | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEEEEGAAAR | EWGATPAGPV | WTAVFDYEAV | GDEELTLRRG | DRVQVLSQDC | AVSGDEGWWT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GQLPSGRVGV | FPSNYVAPAA | PAAPSDLQLP | QEIPFHELQL | EEIIGVGGFG | KVYRAVWRGE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EVAVKAARLD | PERDPAVTAE | QVRQEARLFG | ALQHPNIIAL | RGACLSPPNL | CLVMEYARGG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ALSRVLAGRR | VPPHVLVNWA | VQVARGMNYL | HNDAPVPIIH | RDLKSINILI | LEAIENHNLA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DTVLKITDFG | LAREWHKTTK | MSAAGTYAWM | APEVIRLSLF | SKSSDVWSFG | VLLWELLTGE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VPYREIDALA | VAYGVAMNKL | TLPIPSTCPE | PFARLLEECW | DPDPHGRPDF | GSILKQLEVI |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EQSALFQMPL | ESFHSLQEDW | KLEIQHMFDD | LRTKEKELRS | REEELLRAAQ | EQRFQEEQLR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RREQELAERE | MDIVERELHL | LMSQLSQEKP | RVRKRKGNFK | RSRLLKLREG | SSHISLPSGF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EHKITVQASP | TLDKRKGSDG | ASPPASPSII | PRLRAIRLTP | MDCGGSSGSG | TWSRSGPPKK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EELVGGKKKG | RTWGPSSTLQ | KERAGGEERL | KALGEGSKQW | SSSAPNLGKS | PKHTPMAPGF |
| 610 | 620 | 630 | 640 | 650 | 660 |
| ASLNEMEEFA | EADEGNNVPP | SPYSTPSYLK | VPLPAEPSPC | VQAPWEPPAV | TPSRPGHGAR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RRCDLALLSC | ATLLSAVGLG | ADVAEARAGD | GEEQRRWLDS | LFFPRPGRFP | RGLSPTGRPG |
| 730 | 740 | 750 | 760 | 770 | 780 |
| GRREDTAPGL | GLAPSATLVS | LSSVSDCNST | RSLLRSDSDE | AAPAAPSPPP | SPLAPSPSTN |
| 790 | 800 | 810 | 820 | 830 | 840 |
| PLVDVELESF | KKDPRQSLTP | THVTAAHAVS | RGHRRTPSDG | ALRQREPLEL | TNHGPRDPLD |
| 850 | 860 | 870 | 880 | 890 | 900 |
| FPRLPDPQAL | FPTRRRPLEF | PGRPTTLTFA | PRPRPAASRP | RLDPWKLVSF | GRTLSISPPS |
| 910 | 920 | 930 | |||
| RPDTPESPGP | PSVQPTLLDM | DMEGQSQDNT | VPLCGVYGSH |