Q64010
Gene name |
Crk (Crko) |
Protein name |
Adapter molecule crk |
Names |
Proto-oncogene c-Crk, p38 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:12928 |
EC number |
|
Protein Class |
SH2-SH3 ADAPTOR PROTEIN-RELATED (PTHR19969) |
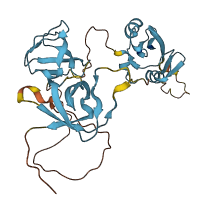
Descriptions
CRK regulates transcription and cytoskeletal reorganization for cell growth and motility by linking tyrosine kinases to small G proteins. The mouse c-Crk protein contains The SH2 and nSH3 domains are used to recruit their target proteins. The cSH3 negatively regulates the function of nSH3. In human Crk, nSH3 domain is also regulated by SH2 domain and inter-SH3 domain region.
Autoinhibitory domains (AIDs)
Target domain |
135-189 (N-terminal Src Homology 3 domain of Ct10 Regulator of Kinase adaptor proteins) |
Relief mechanism |
PTM |
Assay |
Deletion assay |
Target domain |
135-189 (N-terminal Src Homology 3 domain of Ct10 Regulator of Kinase adaptor proteins) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
135-189 (N-terminal Src Homology 3 domain of Ct10 Regulator of Kinase adaptor proteins) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Kobashigawa Y et al. (2007) "Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK", Nature structural & molecular biology, 14, 503-10
- Ogawa S et al. (1994) "The C-terminal SH3 domain of the mouse c-Crk protein negatively regulates tyrosine-phosphorylation of Crk associated p130 in rat 3Y1 cells", Oncogene, 9, 1669-78
- Sriram G et al. (2012) "Commentary: The carboxyl-terminal Crk SH3 domain: Regulatory strategies and new perspectives", FEBS letters, 586, 2615-8
Autoinhibited structure

Activated structure

13 structures for Q64010
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1B07 | X-ray | 250 A | A | 134-190 | PDB |
| 1CKA | X-ray | 150 A | A | 134-190 | PDB |
| 1CKB | X-ray | 190 A | A | 134-190 | PDB |
| 1JU5 | NMR | - | B | 217-228 | PDB |
| 1M30 | NMR | - | A | 134-191 | PDB |
| 1M3A | NMR | - | A | 135-191 | PDB |
| 1M3B | NMR | - | A | 134-191 | PDB |
| 1M3C | NMR | - | A | 132-191 | PDB |
| 2GGR | NMR | - | A | 230-304 | PDB |
| 5IH2 | X-ray | 180 A | A/B | 134-191 | PDB |
| 5JN0 | X-ray | 168 A | A | 6-121 | PDB |
| 5L23 | X-ray | 177 A | A | 134-191 | PDB |
| AF-Q64010-F1 | Predicted | AlphaFoldDB |
No variants for Q64010
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q64010 | |||||
No associated diseases with Q64010
5 regional properties for Q64010
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 11 - 118 | IPR000980 |
| domain | SH3 domain | 132 - 192 | IPR001452-1 |
| domain | SH3 domain | 235 - 296 | IPR001452-2 |
| domain | CRK, N-terminal SH3 domain | 135 - 189 | IPR035457 |
| domain | CRK, C-terminal SH3 domain | 237 - 293 | IPR035458 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR19969 | SH2-SH3 ADAPTOR PROTEIN-RELATED |
| PANTHER Subfamily | PTHR19969:SF8 | ADAPTER MOLECULE CRK |
| PANTHER Protein Class | scaffold/adaptor protein | |
| PANTHER Pathway Category |
Integrin signalling pathway Crk CCKR signaling map CRK Angiogenesis Crk |
|
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
18 GO annotations of molecular function
| Name | Definition |
|---|---|
| cytoskeletal protein binding | Binding to a protein component of a cytoskeleton (actin, microtubule, or intermediate filament cytoskeleton). |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| ephrin receptor binding | Binding to an ephrin receptor. |
| insulin-like growth factor receptor binding | Binding to an insulin-like growth factor receptor. |
| kinase binding | Binding to a kinase, any enzyme that catalyzes the transfer of a phosphate group. |
| phosphotyrosine residue binding | Binding to a phosphorylated tyrosine residue within a protein. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein phosphorylated amino acid binding | Binding to a phosphorylated amino acid residue within a protein. |
| protein self-association | Binding to a domain within the same polypeptide. |
| protein tyrosine kinase binding | Binding to protein tyrosine kinase. |
| protein-macromolecule adaptor activity | The binding activity of a protein that brings together two or more macromolecules in contact, permitting those molecules to function in a coordinated way. The adaptor can bring together two proteins, or a protein and another macromolecule such as a lipid or a nucleic acid. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| scaffold protein binding | Binding to a scaffold protein. Scaffold proteins are crucial regulators of many key signaling pathways. Although not strictly defined in function, they are known to interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. |
| SH2 domain binding | Binding to a SH2 domain (Src homology 2) of a protein, a protein domain of about 100 amino-acid residues and belonging to the alpha + beta domain class. |
| SH3 domain binding | Binding to a SH3 domain (Src homology 3) of a protein, small protein modules containing approximately 50 amino acid residues found in a great variety of intracellular or membrane-associated proteins. |
| signaling adaptor activity | The binding activity of a molecule that brings together two or more molecules in a signaling pathway, permitting those molecules to function in a coordinated way. Adaptor molecules themselves do not have catalytic activity. |
| signaling receptor complex adaptor activity | The binding activity of a molecule that provides a physical support for the assembly of a multiprotein receptor signaling complex. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
42 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| activation of GTPase activity | Any process that initiates the activity of an inactive GTPase through the replacement of GDP by GTP. |
| cell chemotaxis | The directed movement of a motile cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cellular response to endothelin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an endothelin stimulus. Endothelin is any of three secretory vasoconstrictive peptides (endothelin-1, -2, -3). |
| cellular response to insulin-like growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin-like growth factor stimulus. |
| cellular response to nerve growth factor stimulus | A process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nerve growth factor stimulus. |
| cellular response to nitric oxide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nitric oxide stimulus. |
| cellular response to transforming growth factor beta stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a transforming growth factor beta stimulus. |
| cerebellar neuron development | The process whose specific outcome is the progression of a cerebellar neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell. |
| cerebral cortex development | The progression of the cerebral cortex over time from its initial formation until its mature state. The cerebral cortex is the outer layered region of the telencephalon. |
| dendrite development | The process whose specific outcome is the progression of the dendrite over time, from its formation to the mature structure. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| establishment of cell polarity | The specification and formation of anisotropic intracellular organization or cell growth patterns. |
| helper T cell diapedesis | The passage of a helper T cell between the tight junctions of endothelial cells lining blood vessels, typically the fourth and final step of cellular extravasation. |
| hippocampus development | The progression of the hippocampus over time from its initial formation until its mature state. |
| lipid metabolic process | The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids. |
| negative regulation of cell motility | Any process that stops, prevents, or reduces the frequency, rate or extent of cell motility. |
| negative regulation of natural killer cell mediated cytotoxicity | Any process that stops, prevents, or reduces the rate of natural killer mediated cytotoxicity. |
| negative regulation of wound healing | Any process that decreases the rate, frequency, or extent of the series of events that restore integrity to a damaged tissue, following an injury. |
| neuron migration | The characteristic movement of an immature neuron from germinal zones to specific positions where they will reside as they mature. |
| positive regulation of cell growth | Any process that activates or increases the frequency, rate, extent or direction of cell growth. |
| positive regulation of smooth muscle cell migration | Any process that activates, maintains or increases the frequency, rate or extent of smooth muscle cell migration. |
| positive regulation of substrate adhesion-dependent cell spreading | Any process that activates or increases the frequency, rate or extent of substrate adhesion-dependent cell spreading. |
| reelin-mediated signaling pathway | The series of molecular signals initiated by the binding of reelin (a secreted glycoprotein) to a receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell adhesion mediated by integrin | Any process that modulates the frequency, rate, or extent of cell adhesion mediated by integrin. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of dendrite development | Any process that modulates the frequency, rate or extent of dendrite development. |
| regulation of GTPase activity | Any process that modulates the rate of GTP hydrolysis by a GTPase. |
| regulation of intracellular signal transduction | Any process that modulates the frequency, rate or extent of intracellular signal transduction. |
| regulation of leukocyte migration | Any process that modulates the frequency, rate, or extent of leukocyte migration. |
| regulation of protein binding | Any process that modulates the frequency, rate or extent of protein binding. |
| regulation of Rac protein signal transduction | Any process that modulates the frequency, rate or extent of Rac protein signal transduction. |
| regulation of signal transduction | Any process that modulates the frequency, rate or extent of signal transduction. |
| regulation of T cell migration | Any process that modulates the frequency, rate or extent of T cell migration. |
| response to cholecystokinin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cholecystokinin stimulus. |
| response to hepatocyte growth factor | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hepatocyte growth factor stimulus. |
| response to hydrogen peroxide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen peroxide (H2O2) stimulus. |
| response to peptide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a peptide stimulus. |
| response to yeast | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a yeast species. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q04929 | CRK | Adapter molecule crk | Gallus gallus (Chicken) | SS |
| Q9XYM0 | Crk | Adapter molecule Crk | Drosophila melanogaster (Fruit fly) | SS |
| P46109 | CRKL | Crk-like protein | Homo sapiens (Human) | SS |
| P46108 | CRK | Adapter molecule crk | Homo sapiens (Human) | EV |
| O55033 | Nck2 | Cytoplasmic protein NCK2 | Mus musculus (Mouse) | SS |
| Q99M51 | Nck1 | Cytoplasmic protein NCK1 | Mus musculus (Mouse) | PR |
| P47941 | Crkl | Crk-like protein | Mus musculus (Mouse) | SS |
| Q5U2U2 | Crkl | Crk-like protein | Rattus norvegicus (Rat) | SS |
| Q63768 | Crk | Adapter molecule crk | Rattus norvegicus (Rat) | SS |
| Q9NHC3 | ced-2 | Cell death abnormality protein 2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGNFDSEER | SSWYWGRLSR | QEAVALLQGQ | RHGVFLVRDS | STSPGDYVLS | VSENSRVSHY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IINSSGPRPP | VPPSPAQPPP | GVSPSRLRIG | DQEFDSLPAL | LEFYKIHYLD | TTTLIEPVAR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SRQGSGVILR | QEEAEYVRAL | FDFNGNDEED | LPFKKGDILR | IRDKPEEQWW | NAEDSEGKRG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MIPVPYVEKY | RPASASVSAL | IGGNQEGSHP | QPLGGPEPGP | YAQPSVNTPL | PNLQNGPIYA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RVIQKRVPNA | YDKTALALEV | GELVKVTKIN | VSGQWEGECN | GKRGHFPFTH | VRLLDQQNPD |
| EDFS |