Q62689
Gene name |
Jak2 |
Protein name |
Tyrosine-protein kinase JAK2 |
Names |
Janus kinase 2, JAK-2 |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:24514 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
|
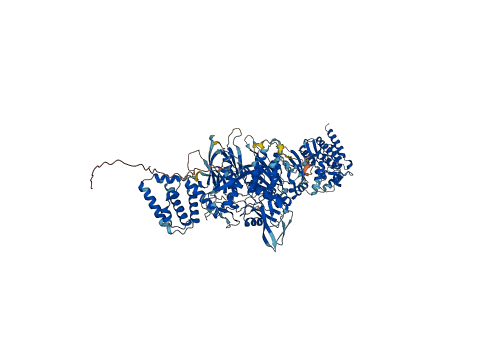
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
849-1126 (Protein kinase domain) |
Relief mechanism |
Partner binding, PTM |
Assay |
|
Accessory elements
698-717 (Activation loop from InterPro)
Target domain |
545-809 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
993-1019 (Activation loop from InterPro)
Target domain |
849-1126 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Ma W et al. (2009) "Mutation profile of JAK2 transcripts in patients with chronic myeloproliferative neoplasias", The Journal of molecular diagnostics : JMD, 11, 49-53
- Saharinen P et al. (2003) "Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain", Molecular biology of the cell, 14, 1448-59
- Wang Q et al. (2010) "Multicolor monitoring of dysregulated protein kinases in chronic myelogenous leukemia", ACS chemical biology, 5, 887-95
- Sotirellis N et al. (1995) "Autophosphorylation induces autoactivation and a decrease in the Src homology 2 domain accessibility of the Lyn protein kinase", The Journal of biological chemistry, 270, 29773-80
- Williams NK et al. (2009) "Crystal structures of the Lyn protein tyrosine kinase domain in its Apo- and inhibitor-bound state", The Journal of biological chemistry, 284, 284-291
- Brian BF 4th et al. (2022) "SH3-domain mutations selectively disrupt Csk homodimerization or PTPN22 binding", Scientific reports, 12, 5875
- Boggon TJ et al. (2004) "Structure and regulation of Src family kinases", Oncogene, 23, 7918-27
- Register AC et al. (2014) "SH2-catalytic domain linker heterogeneity influences allosteric coupling across the SFK family", Biochemistry, 53, 6910-23
- Engen JR et al. (2008) "Structure and dynamic regulation of Src-family kinases", Cellular and molecular life sciences : CMLS, 65, 3058-73
- Alvarado JJ et al. (2010) "Crystal structure of the Src family kinase Hck SH3-SH2 linker regulatory region supports an SH3-dominant activation mechanism", The Journal of biological chemistry, 285, 35455-61
- Hong E et al. (2004) "Solution structure and backbone dynamics of the non-receptor protein-tyrosine kinase-6 Src homology 2 domain", The Journal of biological chemistry, 279, 29700-8
- Ko S et al. (2009) "Structural basis of the auto-inhibition mechanism of nonreceptor tyrosine kinase PTK6", Biochemical and biophysical research communications, 384, 236-42
- Qiu H et al. (2002) "Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition", The Journal of biological chemistry, 277, 34634-41
- Laham LE et al. (2000) "The activation loop in Lck regulates oncogenic potential by inhibiting basal kinase activity and restricting substrate specificity", Oncogene, 19, 3961-70
- Furlan G et al. (2014) "Phosphatase CD45 both positively and negatively regulates T cell receptor phosphorylation in reconstituted membrane protein clusters", The Journal of biological chemistry, 289, 28514-25
- Williams JC et al. (1997) "The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions", Journal of molecular biology, 274, 757-75
- Meng Y et al. (2014) "Locking the active conformation of c-Src kinase through the phosphorylation of the activation loop", Journal of molecular biology, 426, 423-35
Autoinhibited structure

Activated structure

1 structures for Q62689
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q62689-F1 | Predicted | AlphaFoldDB |
4 variants for Q62689
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs197251929 | 469 | R>T | No | Ensembl | |
| rs8174948 | 495 | F>L | No | Ensembl | |
| rs8174946 | 699 | D>E | No | Ensembl | |
| rs199203370 | 724 | I>T | No | Ensembl |
No associated diseases with Q62689
17 regional properties for Q62689
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FERM domain | 37 - 380 | IPR000299 |
| domain | Protein kinase domain | 545 - 809 | IPR000719-1 |
| domain | Protein kinase domain | 849 - 1126 | IPR000719-2 |
| domain | SH2 domain | 397 - 487 | IPR000980 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 546 - 805 | IPR001245-1 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 849 - 1120 | IPR001245-2 |
| active_site | Tyrosine-protein kinase, active site | 972 - 984 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 855 - 883 | IPR017441 |
| domain | FERM central domain | 152 - 261 | IPR019748 |
| domain | Band 4.1 domain | 33 - 270 | IPR019749 |
| domain | Janus kinase 2, pseudokinase domain | 545 - 806 | IPR035588 |
| domain | Janus kinase 2, catalytic domain | 844 - 1127 | IPR035589 |
| domain | Tyrosine-protein kinase JAK2, SH2 domain | 386 - 482 | IPR035860 |
| domain | JAK2, FERM domain C-lobe | 266 - 386 | IPR037838 |
| domain | JAK, FERM F2 lobe domain | 143 - 261 | IPR041046 |
| domain | FERM F1 lobe ubiquitin-like domain | 39 - 134 | IPR041155 |
| domain | JAK1-3/TYK2, pleckstrin homology-like domain | 302 - 381 | IPR041381 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
14 GO annotations of cellular component
| Name | Definition |
|---|---|
| caveola | A membrane raft that forms small pit, depression, or invagination that communicates with the outside of a cell and extends inward, indenting the cytoplasm and the cell membrane. Examples include flask-shaped invaginations of the plasma membrane in adipocytes associated with caveolin proteins, and minute pits or incuppings of the cell membrane formed during pinocytosis. Caveolae may be pinched off to form free vesicles within the cytoplasm. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endomembrane system | A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles. |
| euchromatin | A dispersed and relatively uncompacted form of chromatin that is in a transcription-competent conformation. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| granulocyte macrophage colony-stimulating factor receptor complex | The heterodimeric receptor for granulocyte macrophage colony-stimulating factor. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| postsynapse | The part of a synapse that is part of the post-synaptic cell. |
20 GO annotations of molecular function
| Name | Definition |
|---|---|
| acetylcholine receptor binding | Binding to an acetylcholine receptor. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| growth hormone receptor binding | Binding to a growth hormone receptor. |
| heme binding | Binding to a heme, a compound composed of iron complexed in a porphyrin (tetrapyrrole) ring. |
| histone binding | Binding to a histone, any of a group of water-soluble proteins found in association with the DNA of eukaryotic or archaeal chromosomes. They are involved in the condensation and coiling of chromosomes during cell division and have also been implicated in gene regulation and DNA replication. They may be chemically modified (methylated, acetlyated and others) to regulate gene transcription. |
| histone kinase activity (H3-Y41 specific) | Catalysis of the transfer of a phosphate group to the tyrosine-41 residue of histone H3. |
| identical protein binding | Binding to an identical protein or proteins. |
| insulin receptor substrate binding | Binding to an insulin receptor substrate (IRS) protein, an adaptor protein that bind to the transphosphorylated insulin and insulin-like growth factor receptors, are themselves phosphorylated and in turn recruit SH2 domain-containing signaling molecules to form a productive signaling complex. |
| interleukin-12 receptor binding | Binding to an interleukin-12 receptor. |
| metal ion binding | Binding to a metal ion. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction: ATP + protein L-tyrosine = ADP + protein L-tyrosine phosphate by a non-membrane spanning protein. |
| peptide hormone receptor binding | Binding to a receptor for a peptide hormone. |
| phosphatidylinositol 3-kinase binding | Binding to a phosphatidylinositol 3-kinase, any enzyme that catalyzes the addition of a phosphate group to an inositol lipid at the 3' position of the inositol ring. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| SH2 domain binding | Binding to a SH2 domain (Src homology 2) of a protein, a protein domain of about 100 amino-acid residues and belonging to the alpha + beta domain class. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| type 1 angiotensin receptor binding | Binding to a type 1 angiotensin receptor. |
95 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that initiates the activity of the inactive enzyme cysteine-type endopeptidase in the context of an apoptotic process. |
| activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | Any process that initiates the activity of an inactive cysteine-type endopeptidase involved in the apoptotic signaling pathway. |
| activation of Janus kinase activity | The process of introducing a phosphate group to a tyrosine residue of a JAK (Janus Activated Kinase) protein, thereby activating it. |
| adaptive immune response | An immune response mediated by cells expressing specific receptors for antigen produced through a somatic diversification process, and allowing for an enhanced secondary response to subsequent exposures to the same antigen (immunological memory). |
| axon regeneration | The regrowth of axons following their loss or damage. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cellular response to dexamethasone stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a dexamethasone stimulus. |
| cellular response to interleukin-3 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-3 stimulus. |
| cellular response to lipopolysaccharide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| chromatin organization | The assembly or remodeling of chromatin composed of DNA complexed with histones, other associated proteins, and sometimes RNA. |
| collagen-activated signaling pathway | The series of molecular signals initiated by collagen binding to a cell surface receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| cytokine-mediated signaling pathway | The series of molecular signals initiated by the binding of a cytokine to a receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| enzyme linked receptor protein signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell, where the receptor possesses catalytic activity or is closely associated with an enzyme such as a protein kinase, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| erythrocyte differentiation | The process in which a myeloid precursor cell acquires specializes features of an erythrocyte. |
| extrinsic apoptotic signaling pathway | The series of molecular signals in which a signal is conveyed from the cell surface to trigger the apoptotic death of a cell. The pathway starts with either a ligand binding to a cell surface receptor, or a ligand being withdrawn from a cell surface receptor (e.g. in the case of signaling by dependence receptors), and ends when the execution phase of apoptosis is triggered. |
| G protein-coupled receptor signaling pathway | The series of molecular signals initiated by a ligand binding to its receptor, in which the activated receptor promotes the exchange of GDP for GTP on the alpha-subunit of an associated heterotrimeric G-protein complex. The GTP-bound activated alpha-G-protein then dissociates from the beta- and gamma-subunits to further transmit the signal within the cell. The pathway begins with receptor-ligand interaction, and ends with regulation of a downstream cellular process. The pathway can start from the plasma membrane, Golgi or nuclear membrane. |
| granulocyte-macrophage colony-stimulating factor signaling pathway | The series of molecular signals initiated by the binding of the cytokine granulocyte macrophage colony-stimulating factor (GM-CSF) to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. GM-CSF binds to a heterodimer receptor (CSF2R) consisting of an alpha ligand-binding subunit, and a common beta subunit that is shared with other cytokine receptors. |
| growth hormone receptor signaling pathway | The series of molecular signals generated as a consequence of growth hormone receptor binding to its physiological ligand. |
| growth hormone receptor signaling pathway via JAK-STAT | The process in which STAT proteins (Signal Transducers and Activators of Transcription) are activated by members of the JAK (janus activated kinase) family of tyrosine kinases, following the binding of physiological ligands to the growth hormone receptor. Once activated, STATs dimerize and translocate to the nucleus and modulate the expression of target genes. |
| histone H3-Y41 phosphorylation | OBSOLETE. The modification of histone H3 by the addition of a phosphate group to a tyrosine residue at position 41 of the histone. |
| hormone-mediated signaling pathway | The series of molecular signals mediated by the detection of a hormone. |
| interferon-gamma-mediated signaling pathway | The series of molecular signals initiated by interferon-gamma binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. Interferon gamma is the only member of the type II interferon found so far. |
| interleukin-12-mediated signaling pathway | The series of molecular signals initiated by interleukin-12 binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| intrinsic apoptotic signaling pathway in response to oxidative stress | The series of molecular signals in which an intracellular signal is conveyed to trigger the apoptotic death of a cell. The pathway is induced in response to oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, and ends when the execution phase of apoptosis is triggered. |
| mammary gland epithelium development | The process whose specific outcome is the progression of the mammary gland epithelium over time, from its formation to the mature structure. The mammary gland is a large compound sebaceous gland that in female mammals is modified to secrete milk. |
| microglial cell activation | The change in morphology and behavior of a microglial cell resulting from exposure to a cytokine, chemokine, cellular ligand, or soluble factor. |
| mineralocorticoid receptor signaling pathway | The series of molecular signals initiated by mineralocorticoid binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| modulation of chemical synaptic transmission | Any process that modulates the frequency or amplitude of synaptic transmission, the process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse. Amplitude, in this case, refers to the change in postsynaptic membrane potential due to a single instance of synaptic transmission. |
| myeloid cell differentiation | The process in which a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte, megakaryocyte, thrombocyte, or erythrocyte lineages. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of cardiac muscle cell apoptotic process | Any process that decreases the rate or extent of cardiac cell apoptotic process, a form of programmed cell death induced by external or internal signals that trigger the activity of proteolytic caspases whose actions dismantle a cardiac muscle cell and result in its death. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of cell-cell adhesion | Any process that stops, prevents or reduces the rate or extent of cell adhesion to another cell. |
| negative regulation of DNA binding | Any process that stops or reduces the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| negative regulation of heart contraction | Any process that stops, prevents, or reduces the frequency, rate or extent of heart contraction. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| platelet-derived growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a platelet-derived growth factor receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of apoptotic signaling pathway. |
| positive regulation of cell activation | Any process that activates or increases the frequency, rate or extent of activation. |
| positive regulation of cell differentiation | Any process that activates or increases the frequency, rate or extent of cell differentiation. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of cell-substrate adhesion | Any process that increases the frequency, rate or extent of cell-substrate adhesion. Cell-substrate adhesion is the attachment of a cell to the underlying substrate via adhesion molecules. |
| positive regulation of cold-induced thermogenesis | Any process that activates or increases the frequency, rate or extent of cold-induced thermogenesis. |
| positive regulation of cytosolic calcium ion concentration | Any process that increases the concentration of calcium ions in the cytosol. |
| positive regulation of DNA binding | Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of DNA-binding transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| positive regulation of epithelial cell apoptotic process | Any process that activates or increases the frequency, rate or extent of epithelial cell apoptotic process. |
| positive regulation of growth factor dependent skeletal muscle satellite cell proliferation | Any process that activates or increases the frequency, rate or extent of satellite cell proliferation; dependent on specific growth factor activity such as fibroblast growth factors and transforming growth factor beta. |
| positive regulation of growth hormone receptor signaling pathway | Any process that increases the rate, frequency or extent of the growth hormone receptor signaling pathway. The growth hormone receptor signaling pathway is the series of molecular signals generated as a consequence of growth hormone receptor binding to its physiological ligand. |
| positive regulation of inflammatory response | Any process that activates or increases the frequency, rate or extent of the inflammatory response. |
| positive regulation of insulin secretion | Any process that activates or increases the frequency, rate or extent of the regulated release of insulin. |
| positive regulation of interferon-gamma production | Any process that activates or increases the frequency, rate, or extent of interferon-gamma production. Interferon-gamma is also known as type II interferon. |
| positive regulation of interleukin-1 beta production | Any process that activates or increases the frequency, rate, or extent of interleukin-1 beta production. |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| positive regulation of MHC class II biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of MHC class II. |
| positive regulation of natural killer cell proliferation | Any process that activates or increases the frequency, rate or extent of natural killer cell proliferation. |
| positive regulation of nitric oxide biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nitric oxide. |
| positive regulation of nitric-oxide synthase biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of a nitric oxide synthase enzyme. |
| positive regulation of NK T cell proliferation | Any process that activates or increases the frequency, rate or extent of natural killer T cell proliferation. |
| positive regulation of peptidyl-tyrosine phosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation of peptidyl-tyrosine. |
| positive regulation of phosphatidylinositol 3-kinase signaling | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the phosphatidylinositol 3-kinase cascade. |
| positive regulation of phosphoprotein phosphatase activity | Any process that activates or increases the activity of a phosphoprotein phosphatase. |
| positive regulation of platelet activation | Any process that increases the rate or frequency of platelet activation. Platelet activation is a series of progressive, overlapping events triggered by exposure of the platelets to subendothelial tissue. |
| positive regulation of platelet aggregation | Any process that activates or increases the frequency, rate or extent of platelet aggregation. Platelet aggregation is the adhesion of one platelet to one or more other platelets via adhesion molecules. |
| positive regulation of protein import into nucleus | Any process that activates or increases the frequency, rate or extent of movement of proteins from the cytoplasm into the nucleus. |
| positive regulation of signaling receptor activity | Any process that activates or increases the frequency, rate or extent of signaling receptor activity. |
| positive regulation of SMAD protein signal transduction | Any process that increases the rate, frequency or extent of SMAD protein signal transduction. Pathway-restricted SMAD proteins and common-partner SMAD proteins are involved in the transforming growth factor beta receptor signaling pathways. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of tumor necrosis factor production | Any process that activates or increases the frequency, rate or extent of tumor necrosis factor production. |
| positive regulation of tyrosine phosphorylation of STAT protein | Any process that activates or increases the frequency, rate or extent of the introduction of a phosphate group to a tyrosine residue of a STAT (Signal Transducer and Activator of Transcription) protein. |
| positive regulation of vascular associated smooth muscle cell proliferation | Any process that activates or increases the frequency, rate or extent of vascular smooth muscle cell proliferation. |
| post-embryonic hemopoiesis | The stages of blood cell formation that take place after completion of embryonic development. |
| postsynapse to nucleus signaling pathway | The series of molecular signals that conveys information from the postsynapse to the nucleus via cytoskeletal transport of a protein from a postsynapse to the component to the nucleus where it affects biochemical processes that occur in the nucleus (e.g DNA transcription, mRNA splicing, or DNA/histone modifications). |
| programmed cell death induced by symbiont | Cell death resulting from activation of endogenous cellular processes after interaction with a symbiont (defined as the smaller of two, or more, organisms engaged in symbiosis, a close interaction encompassing mutualism through parasitism). This can be triggered by direct interaction with the organism, for example, contact with penetrating hyphae of a fungus; or an indirect interaction such as symbiont-secreted molecules. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| receptor signaling pathway via JAK-STAT | Any process in which STAT proteins (Signal Transducers and Activators of Transcription) and JAK (Janus Activated Kinase) proteins convey a signal to trigger a change in the activity or state of a cell. The receptor signaling pathway via JAK-STAT begins with activation of a receptor and proceeeds through STAT protein activation by members of the JAK family of tyrosine kinases. STAT proteins dimerize and subsequently translocate to the nucleus. The pathway ends with regulation of target gene expression by STAT proteins. |
| regulation of apoptotic process | Any process that modulates the occurrence or rate of cell death by apoptotic process. |
| regulation of inflammatory response | Any process that modulates the frequency, rate or extent of the inflammatory response, the immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. |
| regulation of nitric oxide biosynthetic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nitric oxide. |
| response to amine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an amine stimulus. An amine is a compound formally derived from ammonia by replacing one, two or three hydrogen atoms by hydrocarbyl groups. |
| response to antibiotic | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an antibiotic stimulus. An antibiotic is a chemical substance produced by a microorganism which has the capacity to inhibit the growth of or to kill other microorganisms. |
| response to hydroperoxide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydroperoxide stimulus. Hydroperoxides are monosubstitution products of hydrogen peroxide, HOOH. |
| response to interleukin-12 | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-12 stimulus. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| response to organic cyclic compound | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic cyclic compound stimulus. |
| response to oxidative stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| response to tumor necrosis factor | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a tumor necrosis factor stimulus. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| tumor necrosis factor-mediated signaling pathway | The series of molecular signals initiated by tumor necrosis factor binding to its receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| tyrosine phosphorylation of STAT protein | The process of introducing a phosphate group to a tyrosine residue of a STAT (Signal Transducer and Activator of Transcription) protein. |
76 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0JNB0 | FYN | Tyrosine-protein kinase Fyn | Bos taurus (Bovine) | SS |
| Q0VBZ0 | CSK | Tyrosine-protein kinase CSK | Bos taurus (Bovine) | SS |
| Q3ZC95 | BTK | Tyrosine-protein kinase | Bos taurus (Bovine) | EV SS |
| P42683 | LCK | Proto-oncogene tyrosine-protein kinase LCK | Gallus gallus (Chicken) | SS |
| P41239 | CSK | Tyrosine-protein kinase CSK | Gallus gallus (Chicken) | SS |
| P00523 | SRC | Proto-oncogene tyrosine-protein kinase Src | Gallus gallus (Chicken) | EV |
| Q02977 | YRK | Proto-oncogene tyrosine-protein kinase Yrk | Gallus gallus (Chicken) | SS |
| Q8JH64 | BTK | Tyrosine-protein kinase BTK | Gallus gallus (Chicken) | SS |
| P09324 | YES1 | Tyrosine-protein kinase Yes | Gallus gallus (Chicken) | SS |
| Q05876 | FYN | Tyrosine-protein kinase Fyn | Gallus gallus (Chicken) | SS |
| Q75R65 | JAK2 | Tyrosine-protein kinase JAK2 | Gallus gallus (Chicken) | SS |
| P08630 | Btk | Tyrosine-protein kinase Btk | Drosophila melanogaster (Fruit fly) | SS |
| Q9V9J3 | Src42A | Tyrosine-protein kinase Src42A | Drosophila melanogaster (Fruit fly) | SS |
| P00528 | Src64B | Tyrosine-protein kinase Src64B | Drosophila melanogaster (Fruit fly) | SS |
| Q24592 | hop | Tyrosine-protein kinase hopscotch | Drosophila melanogaster (Fruit fly) | PR |
| P41240 | CSK | Tyrosine-protein kinase CSK | Homo sapiens (Human) | SS |
| P51451 | BLK | Tyrosine-protein kinase Blk | Homo sapiens (Human) | SS |
| P06239 | LCK | Tyrosine-protein kinase Lck | Homo sapiens (Human) | EV |
| P23458 | JAK1 | Tyrosine-protein kinase JAK1 | Homo sapiens (Human) | SS |
| P06241 | FYN | Tyrosine-protein kinase Fyn | Homo sapiens (Human) | SS |
| P51813 | BMX | Cytoplasmic tyrosine-protein kinase BMX | Homo sapiens (Human) | SS |
| P12931 | SRC | Proto-oncogene tyrosine-protein kinase Src | Homo sapiens (Human) | EV |
| P09769 | FGR | Tyrosine-protein kinase Fgr | Homo sapiens (Human) | SS |
| P42680 | TEC | Tyrosine-protein kinase Tec | Homo sapiens (Human) | SS |
| P42679 | MATK | Megakaryocyte-associated tyrosine-protein kinase | Homo sapiens (Human) | SS |
| P52333 | JAK3 | Tyrosine-protein kinase JAK3 | Homo sapiens (Human) | SS |
| Q08881 | ITK | Tyrosine-protein kinase ITK/TSK | Homo sapiens (Human) | EV |
| P07948 | LYN | Tyrosine-protein kinase Lyn | Homo sapiens (Human) | SS |
| P29597 | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | Homo sapiens (Human) | EV |
| Q13882 | PTK6 | Protein-tyrosine kinase 6 | Homo sapiens (Human) | EV |
| P08631 | HCK | Tyrosine-protein kinase HCK | Homo sapiens (Human) | EV |
| P07947 | YES1 | Tyrosine-protein kinase Yes | Homo sapiens (Human) | SS |
| P42685 | FRK | Tyrosine-protein kinase FRK | Homo sapiens (Human) | EV |
| Q06187 | BTK | Tyrosine-protein kinase BTK | Homo sapiens (Human) | EV |
| O60674 | JAK2 | Tyrosine-protein kinase JAK2 | Homo sapiens (Human) | EV |
| Q9R117 | Tyk2 | Non-receptor tyrosine-protein kinase TYK2 | Mus musculus (Mouse) | SS |
| P08103 | Hck | Tyrosine-protein kinase HCK | Mus musculus (Mouse) | SS |
| P16277 | Blk | Tyrosine-protein kinase Blk | Mus musculus (Mouse) | SS |
| Q62270 | Srms | Tyrosine-protein kinase Srms | Mus musculus (Mouse) | SS |
| Q64434 | Ptk6 | Protein-tyrosine kinase 6 | Mus musculus (Mouse) | SS |
| P05480 | Src | Proto-oncogene tyrosine-protein kinase Src | Mus musculus (Mouse) | EV |
| P14234 | Fgr | Tyrosine-protein kinase Fgr | Mus musculus (Mouse) | SS |
| P35991 | Btk | Tyrosine-protein kinase BTK | Mus musculus (Mouse) | EV |
| P41241 | Csk | Tyrosine-protein kinase CSK | Mus musculus (Mouse) | EV |
| P25911 | Lyn | Tyrosine-protein kinase Lyn | Mus musculus (Mouse) | EV |
| Q62137 | Jak3 | Tyrosine-protein kinase JAK3 | Mus musculus (Mouse) | SS |
| P06240 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Mus musculus (Mouse) | SS |
| P24604 | Tec | Tyrosine-protein kinase Tec | Mus musculus (Mouse) | SS |
| Q04736 | Yes1 | Tyrosine-protein kinase Yes | Mus musculus (Mouse) | SS |
| P39688 | Fyn | Tyrosine-protein kinase Fyn | Mus musculus (Mouse) | SS |
| P52332 | Jak1 | Tyrosine-protein kinase JAK1 | Mus musculus (Mouse) | SS |
| Q03526 | Itk | Tyrosine-protein kinase ITK/TSK | Mus musculus (Mouse) | SS |
| P41242 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q922K9 | Frk | Tyrosine-protein kinase FRK | Mus musculus (Mouse) | SS |
| Q62120 | Jak2 | Tyrosine-protein kinase JAK2 | Mus musculus (Mouse) | EV |
| A1Y2K1 | FYN | Tyrosine-protein kinase Fyn | Sus scrofa (Pig) | SS |
| O19064 | JAK2 | Tyrosine-protein kinase JAK2 | Sus scrofa (Pig) | SS |
| Q62662 | Frk | Tyrosine-protein kinase FRK | Rattus norvegicus (Rat) | SS |
| Q62844 | Fyn | Tyrosine-protein kinase Fyn | Rattus norvegicus (Rat) | SS |
| Q07014 | Lyn | Tyrosine-protein kinase Lyn | Rattus norvegicus (Rat) | SS |
| P50545 | Hck | Tyrosine-protein kinase HCK | Rattus norvegicus (Rat) | SS |
| Q9WUD9 | Src | Proto-oncogene tyrosine-protein kinase Src | Rattus norvegicus (Rat) | SS |
| Q01621 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Rattus norvegicus (Rat) | SS |
| Q6P6U0 | Fgr | Tyrosine-protein kinase Fgr | Rattus norvegicus (Rat) | SS |
| P32577 | Csk | Tyrosine-protein kinase CSK | Rattus norvegicus (Rat) | SS |
| P41243 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Rattus norvegicus (Rat) | SS |
| F1LM93 | Yes1 | Tyrosine-protein kinase Yes | Rattus norvegicus (Rat) | SS |
| Q63272 | Jak3 | Tyrosine-protein kinase JAK3 | Rattus norvegicus (Rat) | SS |
| O45539 | src-2 | Tyrosine protein-kinase src-2 | Caenorhabditis elegans | SS |
| G5ECJ6 | csk-1 | Tyrosine-protein kinase csk-1 | Caenorhabditis elegans | SS |
| G5EE56 | src-1 | Tyrosine protein-kinase src-1 | Caenorhabditis elegans | SS |
| A1A5H8 | yes1 | Tyrosine-protein kinase yes | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| F1RDG9 | fynb | Tyrosine-protein kinase fynb | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O12990 | jak1 | Tyrosine-protein kinase JAK1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q1JPZ3 | src | Proto-oncogene tyrosine-protein kinase Src | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6EWH2 | fyna | Tyrosine-protein kinase fyna | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGMACLTMTE | MEGTSTSPAH | QNGDIPGNAN | SVKQTEPVLQ | VYLYHSLGQA | EGDYLKFPNG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EYVAEEICVA | ASKACGITPV | YHNMFALMSE | TERIWYPPNH | VFHIDESTRH | NILYRIRFYF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PHWYCSGSNR | TYRYGVSRGA | EAPLLDDFVM | SYLFAQWRHD | FVHGWIKVPV | THETQEECLG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MAVLDMMRIA | KEKDQTPLAV | YNSISYKTFL | PKCVRAKIQD | YHILTRKRIR | YRFRRFIQQF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SQCKATARNL | KLKYLINLET | LQSAFYTEQF | EVKESARGPS | GEEIFATIII | TGNGGIQWSR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GKHKESETLT | EQDLQLYCDF | PDIIDVSIKQ | ANQECSTESR | VVTVHKQDGK | VLEIELSSLK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EALSFVSLID | GYYRLTADAH | HYLCKEVAPP | AVLENIHSNC | HGPISMDFAI | SKLKKAGNQT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GLYVLRCSPK | DFNKYFLTFA | VERENVIEYK | HCLITKNENG | EYNLSGTKRN | FSSLKDLLNC |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YQMETVRSDS | IIFQFTKCCP | PKPKDKSNLL | VFRTNGVSDV | QLSPTLQRHN | NVNQMVFHKI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RNEDLIFNES | LGQGTFTKIF | KGVRREVGDY | GQLHETEVLL | KVLDKAHRNY | SESFFEAASM |
| 610 | 620 | 630 | 640 | 650 | 660 |
| MSQLSHKHLV | LNYGVCVCGE | ENILVQEFVK | FGSLDTYLKK | NKNSINILWK | LGVAKQLAWA |
| 670 | 680 | 690 | 700 | 710 | 720 |
| MHFLEEKSLI | HGNVCAKNIL | LIREEDRKTG | NPPFIKLSDP | GISITVLPKD | ILQERIPWVP |
| 730 | 740 | 750 | 760 | 770 | 780 |
| PECIENPKNL | TLATDKWSFG | TTLWEICSGG | DKPLSALDSQ | RKLQFYEDKH | QLPAPKWTEL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| ANLINTCMDY | EPDFRPAFRA | VIRDLNSLFT | PDYELLTEND | MLPNMRIGAL | GFSGAFEDRD |
| 850 | 860 | 870 | 880 | 890 | 900 |
| PTQFEERHLK | FLQQLGKGNF | GSVEMCRYDP | LQDNTGEVVA | VKKLQHSTEE | HLRDFEREIE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| ILKSLQHDNI | VKYKGVCYSA | GRRNLRLIME | YLPYGSLRDY | LQKHKERIDH | KKLLQYTSQI |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| CKGMEYLGTK | RYIHRDLATR | NILVENENRV | KIGDFGLTKV | LPQDKEYYKV | KEPGESPIFW |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| YAPESLTESK | FSVASDVWSF | GVVLYELFTY | IEKSKSPPVE | FMRMIGNDKQ | GQMIVFHLIE |
| 1090 | 1100 | 1110 | 1120 | 1130 | |
| LLKNNGRLPR | PEGCPDEIYV | IMTECWNNNV | NQRPSFRDLS | LRVDQIRDSM | AA |