Q62419
Gene name |
Sh3gl1 (Sh3d2b) |
Protein name |
Endophilin-A2 |
Names |
Endophilin-2, SH3 domain protein 2B, SH3 domain-containing GRB2-like protein 1, SH3p8 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:20405 |
EC number |
|
Protein Class |
|
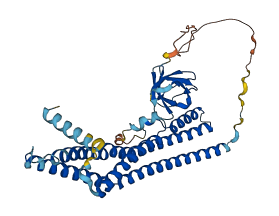
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q62419
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q62419-F1 | Predicted | AlphaFoldDB |
11 variants for Q62419
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389465339 | 23 | G>E | No | EVA | |
| rs3389472228 | 29 | L>P | No | EVA | |
| rs13468388 | 35 | D>E | No | EVA | |
| rs3389471732 | 41 | D>N | No | EVA | |
| rs3389465293 | 44 | S>C | No | EVA | |
| rs3389488767 | 87 | P>A | No | EVA | |
| rs3389471669 | 99 | R>G | No | EVA | |
| rs3411306099 | 117 | A>T | No | EVA | |
| rs3389488783 | 142 | P>S | No | EVA | |
| rs3389451351 | 246 | E>D | No | EVA | |
| rs46615936 | 292 | S>A | No | EVA |
No associated diseases with Q62419
Functions
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| anchoring junction | A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome membrane | The lipid bilayer surrounding an early endosome. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| hippocampal mossy fiber to CA3 synapse | One of the giant synapses that form between the mossy fiber axons of dentate gyrus granule cells and the large complex spines of CA3 pyramidal cells. It consists of a giant bouton known as the mossy fiber expansion, synapsed to the complex, multiheaded spine (thorny excresence) of a CA3 pyramidal cell. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| podosome | An actin-rich adhesion structure characterized by formation upon cell substrate contact and localization at the substrate-attached part of the cell, contain an F-actin-rich core surrounded by a ring structure containing proteins such as vinculin and talin, and have a diameter of 0.5 mm. |
| postsynaptic density, intracellular component | A network of proteins adjacent to the postsynaptic membrane forming an electron dense disc. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize neurotransmitter receptors in the adjacent membrane, such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
| Schaffer collateral - CA1 synapse | A synapse between the Schaffer collateral axon of a CA3 pyramidal cell and a CA1 pyramidal cell. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| beta-1 adrenergic receptor binding | Binding to a beta-1 adrenergic receptor. |
| GTPase binding | Binding to a GTPase, any enzyme that catalyzes the hydrolysis of GTP. |
| identical protein binding | Binding to an identical protein or proteins. |
| lipid binding | Binding to a lipid. |
| phosphatase binding | Binding to a phosphatase. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| SH3 domain binding | Binding to a SH3 domain (Src homology 3) of a protein, small protein modules containing approximately 50 amino acid residues found in a great variety of intracellular or membrane-associated proteins. |
| transmembrane transporter binding | Binding to a transmembrane transporter, a protein or protein complex that enables the transfer of a substance, usually a specific substance or a group of related substances, from one side of a membrane to the other. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| modulation of excitatory postsynaptic potential | Any process that modulates the frequency, rate or extent of excitatory postsynaptic potential (EPSP). EPSP is a process that leads to a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. The flow of ions that causes an EPSP is an excitatory postsynaptic current (EPSC) and makes it easier for the neuron to fire an action potential. |
| positive regulation of synaptic vesicle endocytosis | Any process that activates or increases the frequency, rate or extent of synaptic vesicle endocytosis. |
| regulation of synaptic vesicle endocytosis | Any process that modulates the frequency, rate or extent of synaptic vesicle endocytosis. |
| synaptic vesicle uncoating | The removal of the protein coat on a synaptic vesicle following the pinching step at the end of budding from the presynaptic membrane. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJA1 | SH3GL1 | Endophilin-A2 | Bos taurus (Bovine) | PR |
| Q8AXU9 | SH3GL3 | Endophilin-A3 | Gallus gallus (Chicken) | PR |
| Q8AXV1 | SH3GL2 | Endophilin-A1 | Gallus gallus (Chicken) | PR |
| Q99963 | SH3GL3 | Endophilin-A3 | Homo sapiens (Human) | PR |
| Q99962 | SH3GL2 | Endophilin-A1 | Homo sapiens (Human) | PR |
| Q99961 | SH3GL1 | Endophilin-A2 | Homo sapiens (Human) | PR |
| Q9Y371 | SH3GLB1 | Endophilin-B1 | Homo sapiens (Human) | PR |
| Q62420 | Sh3gl2 | Endophilin-A1 | Mus musculus (Mouse) | PR |
| Q8BZT2 | Sh3rf2 | E3 ubiquitin-protein ligase SH3RF2 | Mus musculus (Mouse) | PR |
| Q9JK48 | Sh3glb1 | Endophilin-B1 | Mus musculus (Mouse) | PR |
| Q62421 | Sh3gl3 | Endophilin-A3 | Mus musculus (Mouse) | PR |
| Q9JLQ0 | Cd2ap | CD2-associated protein | Mus musculus (Mouse) | SS |
| Q8R550 | Sh3kbp1 | SH3 domain-containing kinase-binding protein 1 | Mus musculus (Mouse) | SS |
| O35180 | Sh3gl3 | Endophilin-A3 | Rattus norvegicus (Rat) | PR |
| O35179 | Sh3gl2 | Endophilin-A1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSVAGLKKQF | YKASQLVSEK | VGGAEGTKLD | DDFKDMEKKV | DVTSKAVAEV | LVRTIEYLQP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NPASRAKLTM | LNTVSKIRGQ | VKNPGYPQSE | GLLGECMVRH | GKELGGESNF | GDALLDAGES |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MKRLAEVKDS | LDIEVKQNFI | DPLQNLCDKD | LKEIQHHLKK | LEGRRLDFDY | KKKRQGKIPD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EELRQALEKF | EESKEVAETS | MHNLLETDIE | QVSQLSALVD | AQLDYHRQAV | QILEELADKL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KRRVREASSR | PKREFKPRPR | EPFELGELEQ | PNGGFPCAPA | PKITASSSFR | SSDKPIRMPS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KSMPPLDQPS | CKALYDFEPE | NDGELGFREG | DLITLTNQID | ENWYEGMLHG | QSGFFPLSYV |
| QVLVPLPQ |