Q62261
Gene name |
Sptbn1 (Elf, Spnb-2, Spnb2, Sptb2) |
Protein name |
Spectrin beta chain, non-erythrocytic 1 |
Names |
Beta-II spectrin, Embryonic liver fodrin, Fodrin beta chain |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:20742 |
EC number |
|
Protein Class |
|
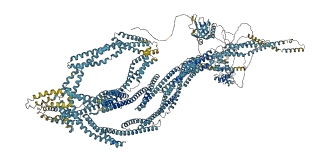
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for Q62261
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1BTN | X-ray | 200 A | A | 2199-2304 | PDB |
| 1MPH | NMR | - | A | 2199-2304 | PDB |
| 6M3P | X-ray | 331 A | A/B | 1591-1910 | PDB |
| AF-Q62261-F1 | Predicted | AlphaFoldDB |
134 variants for Q62261
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389142740 | 22 | N>S | No | EVA | |
| rs3389122360 | 43 | S>T | No | EVA | |
| rs3389140583 | 50 | D>E | No | EVA | |
| rs3402092595 | 97 | G>A | No | EVA | |
| rs3401998282 | 97 | G>R | No | EVA | |
| rs3389110646 | 113 | L>M | No | EVA | |
| rs1133979608 | 209 | N>I | No | EVA | |
| rs3402278517 | 257 | S>R | No | EVA | |
| rs3400804937 | 259 | D>V | No | EVA | |
| rs3389118849 | 297 | A>T | No | EVA | |
| rs3389118821 | 303 | M>I | No | EVA | |
| rs3389142714 | 304 | I>N | No | EVA | |
| rs3389157057 | 308 | E>D | No | EVA | |
| rs3389149777 | 336 | G>E | No | EVA | |
| rs3401430662 | 361 | N>Y | No | EVA | |
| rs3389134530 | 365 | L>P | No | EVA | |
| rs3389140615 | 407 | E>G | No | EVA | |
| rs3389145432 | 421 | E>D | No | EVA | |
| rs3400804959 | 513 | L>F | No | EVA | |
| rs3389110645 | 586 | G>S | No | EVA | |
| rs3389086840 | 603 | P>A | No | EVA | |
| rs3402092641 | 617 | E>A | No | EVA | |
| rs3389134556 | 633 | R>P | No | EVA | |
| rs3389122416 | 703 | D>H | No | EVA | |
| rs3389149821 | 723 | R>L | No | EVA | |
| rs3389153878 | 753 | A>D | No | EVA | |
| rs3389142682 | 754 | D>G | No | EVA | |
| rs3389140641 | 760 | M>L | No | EVA | |
| rs3402728406 | 775 | D>V | No | EVA | |
| rs213662865 | 802 | T>A | No | EVA | |
| rs3389159362 | 818 | P>S | No | EVA | |
| rs3389086819 | 821 | K>T | No | EVA | |
| rs3389146531 | 825 | A>V | No | EVA | |
| rs3389122434 | 863 | L>P | No | EVA | |
| rs3389145451 | 866 | D>N | No | EVA | |
| rs3402010736 | 882 | E>D | No | EVA | |
| rs3401917740 | 885 | E>Q | No | EVA | |
| rs3402017729 | 887 | I>L | No | EVA | |
| rs3389142716 | 890 | R>G | No | EVA | |
| rs3389118786 | 907 | V>M | No | EVA | |
| rs3389149845 | 911 | I>F | No | EVA | |
| rs3389140632 | 929 | Q>K | No | EVA | |
| rs3389142699 | 936 | R>M | No | EVA | |
| rs3389110701 | 949 | D>E | No | EVA | |
| rs3401430681 | 953 | S>A | No | EVA | |
| rs3402157014 | 953 | S>Y | No | EVA | |
| rs3401657430 | 955 | L>Q | No | EVA | |
| rs3410353345 | 956 | S>G | No | EVA | |
| rs3401792935 | 957 | I>S | No | EVA | |
| rs3389159351 | 970 | W>C | No | EVA | |
| rs3389086873 | 973 | E>D | No | EVA | |
| rs256713181 | 982 | Q>H | No | EVA | |
| rs3389110647 | 1004 | D>A | No | EVA | |
| rs3389153827 | 1030 | A>V | No | EVA | |
| rs3402109883 | 1037 | L>P | No | EVA | |
| rs3389145471 | 1085 | I>F | No | EVA | |
| rs3389145470 | 1088 | E>D | No | EVA | |
| rs3389145448 | 1109 | N>H | No | EVA | |
| rs3389140573 | 1109 | N>I | No | EVA | |
| rs3389118838 | 1134 | D>N | No | EVA | |
| rs3389122347 | 1140 | L>P | No | EVA | |
| rs3389149399 | 1156 | K>R | No | EVA | |
| rs3389146506 | 1167 | Q>H | No | EVA | |
| rs3389157046 | 1178 | T>I | No | EVA | |
| rs3389159334 | 1192 | L>F | No | EVA | |
| rs3389086883 | 1192 | L>S | No | EVA | |
| rs3389122383 | 1196 | E>D | No | EVA | |
| rs3389118802 | 1213 | D>E | No | EVA | |
| rs3389149442 | 1217 | T>I | No | EVA | |
| rs3389157005 | 1228 | V>SDG* | No | EVA | |
| rs3389146671 | 1237 | S>N | No | EVA | |
| rs3389157092 | 1247 | Q>R | No | EVA | |
| rs26875745 | 1249 | K>R | No | EVA | |
| rs3389142741 | 1255 | D>N | No | EVA | |
| rs3400804984 | 1269 | M>* | No | EVA | |
| rs3400804958 | 1273 | D>V | No | EVA | |
| rs3402278506 | 1276 | D>H | No | EVA | |
| rs3389149795 | 1303 | Y>C | No | EVA | |
| rs3389159326 | 1337 | G>V | No | EVA | |
| rs3389149822 | 1355 | L>V | No | EVA | |
| rs3389122423 | 1356 | T>N | No | EVA | |
| rs3389153915 | 1361 | M>T | No | EVA | |
| rs3389157008 | 1364 | V>I | No | EVA | |
| rs3389134504 | 1373 | A>V | No | EVA | |
| rs3389122378 | 1381 | K>M | No | EVA | |
| rs3389086824 | 1403 | I>F | No | EVA | |
| rs3389110697 | 1407 | D>N | No | EVA | |
| rs3389086861 | 1433 | K>R | No | EVA | |
| rs3389157076 | 1461 | L>P | No | EVA | |
| rs3389134467 | 1469 | E>V | No | EVA | |
| rs3389110710 | 1475 | S>T | No | EVA | |
| rs3389149837 | 1572 | Q>* | No | EVA | |
| rs3389145497 | 1598 | Y>* | No | EVA | |
| rs3389142768 | 1611 | Q>E | No | EVA | |
| rs3389140556 | 1616 | M>L | No | EVA | |
| rs3412161923 | 1668 | R>H | No | EVA | |
| rs3389146527 | 1698 | H>Y | No | EVA | |
| rs3401998319 | 1709 | D>E | No | EVA | |
| rs3389159390 | 1722 | G>D | No | EVA | |
| rs3389118822 | 1734 | T>M | No | EVA | |
| rs3389122364 | 1818 | F>Y | No | EVA | |
| rs3389157058 | 1832 | L>F | No | EVA | |
| rs13459831 | 1898 | D>G | No | EVA | |
| rs3389142748 | 1908 | V>E | No | EVA | |
| rs3389159371 | 1967 | F>S | No | EVA | |
| rs3389159352 | 2003 | I>T | No | EVA | |
| rs3389145467 | 2012 | W>C | No | EVA | |
| rs3389142688 | 2013 | L>F | No | EVA | |
| rs3389086866 | 2048 | S>* | No | EVA | |
| rs3389157080 | 2050 | D>E | No | EVA | |
| rs3389134524 | 2053 | E>G | No | EVA | |
| rs3389146561 | 2053 | E>K | No | EVA | |
| rs3389145463 | 2073 | F>I | No | EVA | |
| rs3389146530 | 2101 | P>H | No | EVA | |
| rs3389118801 | 2138 | P>L | No | EVA | |
| rs13473735 | 2171 | L>S | No | EVA | |
| rs255747867 | 2172 | D>N | No | EVA | |
| rs242852230 | 2181 | A>S | No | EVA | |
| rs3389157012 | 2182 | Q>E | No | EVA | |
| rs3389146701 | 2201 | G>R | No | EVA | |
| rs3389157013 | 2202 | F>V | No | EVA | |
| rs3389149191 | 2203 | L>P | No | EVA | |
| rs3389140611 | 2207 | H>N | No | EVA | |
| rs3389086849 | 2207 | H>Q | No | EVA | |
| rs3389153831 | 2209 | W>* | No | EVA | |
| rs3389159391 | 2213 | N>D | No | EVA | |
| rs3389147914 | 2216 | A>T | No | EVA | |
| rs3389145483 | 2219 | R>S | No | EVA | |
| rs3402070452 | 2224 | V>I | No | EVA | |
| rs3402277692 | 2225 | Y>H | No | EVA | |
| rs3401394566 | 2227 | V>A | No | EVA | |
| rs3389146689 | 2237 | K>E | No | EVA | |
| rs3389146730 | 2259 | A>S | No | EVA | |
| rs3389086811 | 2282 | N>Y | No | EVA |
No associated diseases with Q62261
36 regional properties for Q62261
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Actinin-type actin-binding domain, conserved site | 56 - 65 | IPR001589-1 |
| conserved_site | Actinin-type actin-binding domain, conserved site | 130 - 154 | IPR001589-2 |
| domain | Pleckstrin homology domain, spectrin-type | 2199 - 2218 | IPR001605-1 |
| domain | Pleckstrin homology domain, spectrin-type | 2219 - 2240 | IPR001605-2 |
| domain | Pleckstrin homology domain, spectrin-type | 2261 - 2278 | IPR001605-3 |
| domain | Pleckstrin homology domain, spectrin-type | 2281 - 2299 | IPR001605-4 |
| domain | Calponin homology domain | 54 - 158 | IPR001715-1 |
| domain | Calponin homology domain | 173 - 278 | IPR001715-2 |
| domain | Pleckstrin homology domain | 2196 - 2308 | IPR001849 |
| repeat | Spectrin repeat | 303 - 411 | IPR002017-1 |
| repeat | Spectrin repeat | 423 - 525 | IPR002017-2 |
| repeat | Spectrin repeat | 530 - 636 | IPR002017-3 |
| repeat | Spectrin repeat | 639 - 742 | IPR002017-4 |
| repeat | Spectrin repeat | 745 - 847 | IPR002017-5 |
| repeat | Spectrin repeat | 850 - 952 | IPR002017-6 |
| repeat | Spectrin repeat | 957 - 1060 | IPR002017-7 |
| repeat | Spectrin repeat | 1063 - 1166 | IPR002017-8 |
| repeat | Spectrin repeat | 1170 - 1259 | IPR002017-9 |
| repeat | Spectrin repeat | 1276 - 1376 | IPR002017-10 |
| repeat | Spectrin repeat | 1381 - 1482 | IPR002017-11 |
| repeat | Spectrin repeat | 1486 - 1590 | IPR002017-12 |
| repeat | Spectrin repeat | 1592 - 1696 | IPR002017-13 |
| repeat | Spectrin repeat | 1698 - 1801 | IPR002017-14 |
| repeat | Spectrin repeat | 1805 - 1907 | IPR002017-15 |
| repeat | Spectrin repeat | 1914 - 2014 | IPR002017-16 |
| repeat | Spectrin repeat | 2018 - 2097 | IPR002017-17 |
| repeat | Spectrin/alpha-actinin | 305 - 411 | IPR018159-1 |
| repeat | Spectrin/alpha-actinin | 425 - 525 | IPR018159-2 |
| repeat | Spectrin/alpha-actinin | 530 - 849 | IPR018159-3 |
| repeat | Spectrin/alpha-actinin | 851 - 1169 | IPR018159-4 |
| repeat | Spectrin/alpha-actinin | 1171 - 1380 | IPR018159-5 |
| repeat | Spectrin/alpha-actinin | 1381 - 1592 | IPR018159-6 |
| repeat | Spectrin/alpha-actinin | 1593 - 1805 | IPR018159-7 |
| repeat | Spectrin/alpha-actinin | 1806 - 2027 | IPR018159-8 |
| repeat | Spectrin/alpha-actinin | 2020 - 2162 | IPR018159-9 |
| domain | Pleckstrin homology domain 9 | 2200 - 2302 | IPR041681 |
Functions
19 GO annotations of cellular component
| Name | Definition |
|---|---|
| axolemma | The portion of the plasma membrane surrounding an axon; it is a specialized trilaminar random mosaic of protein molecules floating within a fluid matrix of highly mobile phospholipid molecules, 7-8 nm in thickness. |
| cell junction | A cellular component that forms a specialized region of connection between two or more cells, or between a cell and the extracellular matrix, or between two membrane-bound components of a cell, such as flagella. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cortical actin cytoskeleton | The portion of the actin cytoskeleton, comprising filamentous actin and associated proteins, that lies just beneath the plasma membrane. |
| cortical cytoskeleton | The portion of the cytoskeleton that lies just beneath the plasma membrane. |
| cuticular plate | A dense network of actin filaments found beneath the apical cell surface of hair cells, and into which stereocilia are inserted. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endomembrane system | A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| M band | The midline of aligned thick filaments in a sarcomere; location of specific proteins that link thick filaments. Depending on muscle type the M band consists of different numbers of M lines. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynapse | The part of a synapse that is part of the post-synaptic cell. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| spectrin | Membrane associated dimeric protein (240 and 220 kDa) of erythrocytes. Forms a complex with ankyrin, actin and probably other components of the membrane cytoskeleton, so that there is a mesh of proteins underlying the plasma membrane, potentially restricting the lateral mobility of integral proteins. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| ankyrin binding | Binding to ankyrin, a 200 kDa cytoskeletal protein that attaches other cytoskeletal proteins to integral membrane proteins. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| GTPase binding | Binding to a GTPase, any enzyme that catalyzes the hydrolysis of GTP. |
| phospholipid binding | Binding to a phospholipid, a class of lipids containing phosphoric acid as a mono- or diester. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| structural constituent of cytoskeleton | The action of a molecule that contributes to the structural integrity of a cytoskeletal structure. |
14 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| actin filament capping | The binding of a protein or protein complex to the end of an actin filament, thus preventing the addition, exchange or removal of further actin subunits. |
| central nervous system development | The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain and spinal cord. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| central nervous system formation | The process that gives rise to the central nervous system. This process pertains to the initial formation of a structure from unspecified parts. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| common-partner SMAD protein phosphorylation | The process of introducing a phosphate group on to a common-partner SMAD protein. A common partner SMAD protein binds to pathway-restricted SMAD proteins forming a complex that translocates to the nucleus. |
| Golgi to plasma membrane protein transport | The directed movement of proteins from the Golgi to the plasma membrane in transport vesicles that move from the trans-Golgi network to the plasma membrane. |
| membrane assembly | The aggregation, arrangement and bonding together of a set of components to form a membrane. |
| mitotic cytokinesis | A cell cycle process that results in the division of the cytoplasm of a cell after mitosis, resulting in the separation of the original cell into two daughter cells. |
| plasma membrane organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the plasma membrane. |
| positive regulation of interleukin-2 production | Any process that activates or increases the frequency, rate, or extent of interleukin-2 production. |
| positive regulation of protein localization to plasma membrane | Any process that activates or increases the frequency, rate or extent of protein localization to plasma membrane. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| regulation of protein localization to plasma membrane | Any process that modulates the frequency, rate or extent of protein localization to plasma membrane. |
| regulation of SMAD protein signal transduction | Any process that modulates the rate, frequency or extent of SMAD protein signal transduction. Pathway-restricted SMAD proteins and common-partner SMAD proteins are involved in the transforming growth factor beta receptor signaling pathways. |
9 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O15020 | SPTBN2 | Spectrin beta chain, non-erythrocytic 2 | Homo sapiens (Human) | PR |
| P11277 | SPTB | Spectrin beta chain, erythrocytic | Homo sapiens (Human) | PR |
| Q01082 | SPTBN1 | Spectrin beta chain, non-erythrocytic 1 | Homo sapiens (Human) | PR |
| Q9JI91 | Actn2 | Alpha-actinin-2 | Mus musculus (Mouse) | SS |
| O88990 | Actn3 | Alpha-actinin-3 | Mus musculus (Mouse) | SS |
| P57780 | Actn4 | Alpha-actinin-4 | Mus musculus (Mouse) | SS |
| Q7TPR4 | Actn1 | Alpha-actinin-1 | Mus musculus (Mouse) | SS |
| P15508 | Sptb | Spectrin beta chain, erythrocytic | Mus musculus (Mouse) | PR |
| Q9QWN8 | Sptbn2 | Spectrin beta chain, non-erythrocytic 2 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTTTVATDYD | NIEIQQQYSD | VNNRWDVDDW | DNENSSARLF | ERSRIKALAD | EREAVQKKTF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TKWVNSHLAR | VSCRITDLYT | DLRDGRMLIK | LLEVLSGERL | PKPTKGRMRI | HCLENVDKAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QFLKEQRVHL | ENMGSHDIVD | GNHRLTLGLI | WTIILRFQIQ | DISVETEDNK | EKKSAKDALL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LWCQMKTAGY | PNVNIHNFTT | SWRDGMAFNA | LIHKHRPDLI | DFDKLKKSNA | HYNLQNAFNL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AEQHLGLTKL | LDPEDISVDH | PDEKSIITYV | VTYYHYFSKM | KALAVEGKRI | GKVLDNAIET |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EKMIEKYESL | ASDLLEWIEQ | TIIILNNRKF | ANSLVGVQQQ | LQAFNTYRTV | EKPPKFTEKG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NLEVLLFTIQ | SKMRANNQKV | YMPREGKLIS | DINKAWERLE | KAEHERELAL | RNELIRQEKL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| EQLARRFDRK | AAMRETWLSE | NQRLVSQDNF | GFDLPAVEAA | TKKHEAIETD | IAAYEERVQA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VVAVARELEA | ENYHDIKRIT | ARKDNVIRLW | EYLLELLRAR | RQRLEMNLGL | QKIFQEMLYI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| MDWMDEMKVL | LLSQDYGKHL | LGVEDLLQKH | ALVEADIAIQ | AERVRGVNAS | AQKFATDGEG |
| 610 | 620 | 630 | 640 | 650 | 660 |
| YKPCDPQVIR | DRVAHMEFCY | QELCQLAAER | RARLEESRRL | WKFFWEMAEE | EGWIREKEKI |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LSSDDYGKDL | TSVMRLLSKH | RAFEDEMSGR | SGHFEQAIKE | GEDMIAEEHF | GSEKIRERII |
| 730 | 740 | 750 | 760 | 770 | 780 |
| YIREQWANLE | QLSAIRKKRL | EEASLLHQFQ | ADADDIDAWM | LDILKIVSSN | DVGHDEYSTQ |
| 790 | 800 | 810 | 820 | 830 | 840 |
| SLVKKHKDVA | EEITNYRPTI | DTLHEQASAL | PQAHAESPDV | KGRLAGIEER | CKEMAELTRL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| RKQALQDTLA | LYKMFSEADA | CELWIDEKEQ | WLNNMQIPEK | LEDLEVIQHR | FESLEPEMNN |
| 910 | 920 | 930 | 940 | 950 | 960 |
| QASRVAVVNQ | IARQLMHNGH | PSEKEIRAQQ | DKLNTRWSQF | RELVDRKKDA | LLSALSIQNY |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| HLECNETKSW | IREKTKVIES | TQDLGNDLAG | VMALQRKLTG | MERDLVAIEA | KLSDLQKEAE |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| KLESEHPDQA | QAILSRLAEI | SDVWEEMKTT | LKNREASLGE | ASKLQQFLRD | LDDFQSWLSR |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| TQTAIASEDM | PNTLTEAEKL | LTQHENIKNE | IDNYEEDYQK | MRDMGEMVTQ | GQTDAQYMFL |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| RQRLQALDTG | WNELHKMWEN | RQNLLSQSHA | YQQFLRDTKQ | AEAFLNNQEY | VLAHTEMPTT |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| LEGAEAAIKK | QEDFMTTMDA | NEEKINAVVE | TGRRLVSDGN | INSDRIQEKV | DSIDDRHRKN |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| REAASELLMR | LKDNRDLQKF | LQDCQELSLW | INEKMLTAQD | MSYDEARNLH | SKWLKHQAFM |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| AELASNKEWL | DKIEKEGMQL | ISEKPETEAV | VKEKLTGLHK | MWEVLESTTQ | TKAQRLFDAN |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| KAELFTQSCA | DLDKWLHGLE | SQIQSDDYGK | DLTSVNILLK | KQQMLENQME | VRKKEIEELQ |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| SQAQALSQEG | KSTDEVDSKR | LTVQTKFMEL | LEPLSERKHN | LLASKEIHQF | NRDVEDEILW |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| VGERMPLATS | TDHGHNLQTV | QLLIKKNQTL | QKEIQGHQPR | IDDIFERSQN | IITDSSSLNA |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| EAIRQRLADL | KQLWGLLIEE | TEKRHRRLEE | AHKAQQYYFD | AAEAEAWMSE | QELYMMSEEK |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| AKDEQSAVSM | LKKHQILEQA | VEDYAETVHQ | LSKTSRALVA | DSHPESERIS | MRQSKVDKLY |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| AGLKDLAEER | RGKLDERHRL | FQLNREVDDL | EQWIAEREVV | AGSHELGQDY | EHVTMLQERF |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| REFARDTGNI | GQERVDTVNN | MADELINSGH | SDAATIAEWK | DGLNEAWADL | LELIDTRTQI |
| 1810 | 1820 | 1830 | 1840 | 1850 | 1860 |
| LAASYELHKF | YHDAKEIFGR | IQDKHKKLPE | ELGRDQNTVE | TLQRMHTTFE | HDIQALGTQV |
| 1870 | 1880 | 1890 | 1900 | 1910 | 1920 |
| RQLQEDAARL | QAAYAGDKAD | DIQKRENEVL | EAWKSLLDAC | EGRRVRLVDT | GDKFRFFSMV |
| 1930 | 1940 | 1950 | 1960 | 1970 | 1980 |
| RDLMLWMEDV | IRQIEAQEKP | RDVSSVELLM | NNHQGIKAEI | DARNDSFTAC | IELGKSLLAR |
| 1990 | 2000 | 2010 | 2020 | 2030 | 2040 |
| KHYASEEIKE | KLLQLTEKRK | EMIDKWEDRW | EWLRLILEVH | QFSRDASVAE | AWLLGQEPYL |
| 2050 | 2060 | 2070 | 2080 | 2090 | 2100 |
| SSREIGQSVD | EVEKLIKRHE | AFEKSAATWD | ERFSALERLT | TLELLEVRRQ | QEEEERKRRP |
| 2110 | 2120 | 2130 | 2140 | 2150 | 2160 |
| PSPDPNTKVS | EEAESQQWDT | SKGDQVSQNG | LPAEQGSPRM | AGTMETSEMV | NGAAEQRTSS |
| 2170 | 2180 | 2190 | 2200 | 2210 | 2220 |
| KESSPVPSPT | LDRKAKSALP | AQSAATLPAR | TLETPAAQME | GFLNRKHEWE | AHNKKASSRS |
| 2230 | 2240 | 2250 | 2260 | 2270 | 2280 |
| WHNVYCVINN | QEMGFYKDAK | SAASGIPYHS | EVPVSLKEAI | CEVALDYKKK | KHVFKLRLSD |
| 2290 | 2300 | 2310 | 2320 | 2330 | 2340 |
| GNEYLFQAKD | DEEMNTWIQA | ISSAISSDKH | DTSASTQSTP | ASSRAQTLPT | SVVTITSESS |
| 2350 | 2360 | ||||
| PGKREKDKEK | DKEKRFSLFG | KKK |