Q62190
Gene name |
Mst1r (Ron, Stk) |
Protein name |
Macrophage-stimulating protein receptor |
Names |
MSP receptor , EC 2.7.10.1 , Stem cell-derived tyrosine kinase , p185-Ron , CD antigen CD136 [Cleaved into: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain] |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:19882 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
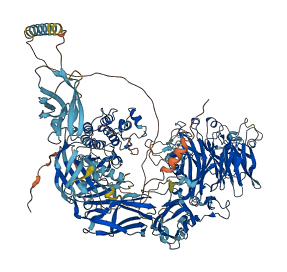
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1059-1322 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
1202-1229 (Activation loop from InterPro)
Target domain |
1059-1322 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
1202-1229 (Activation loop from InterPro)
Target domain |
1059-1322 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Yokoyama N et al. (2005) "The C terminus of RON tyrosine kinase plays an autoinhibitory role", The Journal of biological chemistry, 280, 8893-900
- Huang X et al. (2009) "Structural insights into the inhibited states of the Mer receptor tyrosine kinase", Journal of structural biology, 165, 88-96
- Uchikawa E et al. (2019) "Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex", eLife, 8,
- Nielsen J et al. (2022) "Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling", Journal of molecular biology, 434, 167458
- Chen YS et al. (2021) "Insertion of a synthetic switch into insulin provides metabolite-dependent regulation of hormone-receptor activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Craddock BP et al. (2007) "Autoinhibition of the insulin-like growth factor I receptor by the juxtamembrane region", FEBS letters, 581, 3235-40
Autoinhibited structure

Activated structure

1 structures for Q62190
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q62190-F1 | Predicted | AlphaFoldDB |
75 variants for Q62190
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389081040 | 21 | S>* | No | EVA | |
| rs3412982874 | 42 | F>L | No | EVA | |
| rs3389073758 | 48 | V>I | No | EVA | |
| rs3389073985 | 81 | V>E | No | EVA | |
| rs3389038311 | 92 | L>M | No | EVA | |
| rs3389038280 | 111 | G>* | No | EVA | |
| rs3389016521 | 125 | V>G | No | EVA | |
| rs3400555833 | 131 | P>S | No | EVA | |
| rs3389081038 | 144 | C>Y | No | EVA | |
| rs3389071665 | 161 | P>L | No | EVA | |
| rs3389047551 | 229 | F>S | No | EVA | |
| rs3389016579 | 266 | Q>H | No | EVA | |
| rs3412961647 | 325 | A>T | No | EVA | |
| rs3389052226 | 381 | E>K | No | EVA | |
| rs3389077144 | 453 | L>M | No | EVA | |
| rs3400644776 | 472 | R>L | No | EVA | |
| rs3412775934 | 472 | R>S | No | EVA | |
| rs3389070916 | 476 | V>L | No | EVA | |
| rs3389070916 | 476 | V>M | No | EVA | |
| rs237806376 | 514 | A>T | No | EVA | |
| rs3389047617 | 542 | Q>* | No | EVA | |
| rs3389071641 | 543 | R>L | No | EVA | |
| rs3389064624 | 587 | T>R | No | EVA | |
| rs3389073829 | 613 | V>M | No | EVA | |
| rs3413041980 | 643 | E>V | No | EVA | |
| rs3389061068 | 650 | Q>H | No | EVA | |
| rs3389047566 | 682 | S>F | No | EVA | |
| rs3389052282 | 699 | A>S | No | EVA | |
| rs3389074005 | 702 | T>I | No | EVA | |
| rs3389064642 | 703 | Y>* | No | EVA | |
| rs3389071656 | 719 | V>M | No | EVA | |
| rs261777608 | 723 | G>E | No | EVA | |
| rs229876796 | 725 | Q>R | No | EVA | |
| rs3389038318 | 769 | K>M | No | EVA | |
| rs244300641 | 788 | I>V | No | EVA | |
| rs33276873 | 800 | H>Y | No | EVA | |
| rs227374420 | 801 | F>L | No | EVA | |
| rs3400730560 | 805 | F>C | No | EVA | |
| rs3389071634 | 805 | F>I | No | EVA | |
| rs3413151936 | 805 | F>L | No | EVA | |
| rs246462726 | 806 | H>N | No | EVA | |
| rs246462726 | 806 | H>Y | No | EVA | |
| rs3389070761 | 812 | V>M | No | EVA | |
| rs3389016566 | 862 | R>C | No | EVA | |
| rs3413039604 | 895 | A>P | No | EVA | |
| rs263823196 | 908 | V>I | No | EVA | |
| rs3389038284 | 942 | G>E | No | EVA | |
| rs244409333 | 981 | I>F | No | EVA | |
| rs3389061047 | 986 | R>S | No | EVA | |
| rs227395919 | 999 | T>A | No | EVA | |
| rs3389060998 | 1001 | L>I | No | EVA | |
| rs3400790773 | 1008 | A>S | No | EVA | |
| rs3389038339 | 1011 | A>V | No | EVA | |
| rs33278654 | 1042 | M>T | No | EVA | |
| rs3389081060 | 1046 | E>K | No | EVA | |
| rs3389077203 | 1061 | T>I | No | EVA | |
| rs3389073975 | 1078 | Y>F | No | EVA | |
| rs3389047570 | 1083 | Q>H | No | EVA | |
| rs3389016580 | 1095 | R>H | No | EVA | |
| rs3389052277 | 1098 | E>V | No | EVA | |
| rs3389071691 | 1120 | I>T | No | EVA | |
| rs3389073734 | 1136 | V>M | No | EVA | |
| rs33659070 | 1148 | H>R | No | EVA | |
| rs3389070932 | 1154 | Q>K | No | EVA | |
| rs3413151119 | 1165 | F>I | No | EVA | |
| rs3389071669 | 1199 | V>F | No | EVA | |
| rs3389081052 | 1204 | F>Y | No | EVA | |
| rs219847540 | 1210 | V>I | No | EVA | |
| rs3389068346 | 1215 | Y>* | No | EVA | |
| rs3410608351 | 1227 | P>Q | No | EVA | |
| rs3389070722 | 1228 | V>F | No | EVA | |
| rs3389070722 | 1228 | V>L | No | EVA | |
| rs3389016544 | 1276 | F>I | No | EVA | |
| rs3389081022 | 1282 | R>H | No | EVA | |
| rs3389016546 | 1328 | D>N | No | EVA |
No associated diseases with Q62190
7 regional properties for Q62190
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 680 - 938 | IPR000719 |
| active_site | Serine/threonine-protein kinase, active site | 799 - 811 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 686 - 709 | IPR017441 |
| domain | MAP3K, TRAFs-binding domain | 165 - 545 | IPR025136 |
| domain | MAP3K, PH domain | 556 - 653 | IPR043969 |
| domain | MAP3K, deoxyribohydrolase domain | 115 - 150 | IPR046872 |
| domain | MAP3K, HisK-N-like globin domain | 1042 - 1173 | IPR046873 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| basal plasma membrane | The region of the plasma membrane located at the basal end of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| stress fiber | A contractile actin filament bundle that consists of short actin filaments with alternating polarity, cross-linked by alpha-actinin and possibly other actin bundling proteins, and with myosin present in a periodic distribution along the fiber. |
| vacuole | A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| hepatocyte growth factor receptor activity | Combining with hepatocyte growth factor receptor ligand and transmitting the signal across the plasma membrane to initiate a change in cell activity. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| liver development | The process whose specific outcome is the progression of the liver over time, from its formation to the mature structure. The liver is an exocrine gland which secretes bile and functions in metabolism of protein and carbohydrate and fat, synthesizes substances involved in the clotting of the blood, synthesizes vitamin A, detoxifies poisonous substances, stores glycogen, and breaks down worn-out erythrocytes. |
| nervous system development | The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state. |
| neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| pancreas development | The process whose specific outcome is the progression of the pancreas over time, from its formation to the mature structure. The pancreas is an endoderm derived structure that produces precursors of digestive enzymes and blood glucose regulating hormones. |
| phagocytosis | A vesicle-mediated transport process that results in the engulfment of external particulate material by phagocytes and their delivery to the lysosome. The particles are initially contained within phagocytic vacuoles (phagosomes), which then fuse with primary lysosomes to effect digestion of the particles. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| response to virus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a virus. |
80 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q769I5 | MET | Hepatocyte growth factor receptor | Bos taurus (Bovine) | PR |
| A0M8S8 | MET | Hepatocyte growth factor receptor | Felis catus (Cat) (Felis silvestris catus) | PR |
| P29376 | LTK | Leukocyte tyrosine kinase receptor | Homo sapiens (Human) | SS |
| P14616 | INSRR | Insulin receptor-related protein | Homo sapiens (Human) | SS |
| P08922 | ROS1 | Proto-oncogene tyrosine-protein kinase ROS | Homo sapiens (Human) | SS |
| Q9UM73 | ALK | ALK tyrosine kinase receptor | Homo sapiens (Human) | EV |
| O43157 | PLXNB1 | Plexin-B1 | Homo sapiens (Human) | EV SS |
| Q9HCM2 | PLXNA4 | Plexin-A4 | Homo sapiens (Human) | SS |
| O75051 | PLXNA2 | Plexin-A2 | Homo sapiens (Human) | SS |
| P51805 | PLXNA3 | Plexin-A3 | Homo sapiens (Human) | SS |
| Q9UIW2 | PLXNA1 | Plexin-A1 | Homo sapiens (Human) | EV SS |
| O15031 | PLXNB2 | Plexin-B2 | Homo sapiens (Human) | SS |
| Q9ULL4 | PLXNB3 | Plexin-B3 | Homo sapiens (Human) | SS |
| P08581 | MET | Hepatocyte growth factor receptor | Homo sapiens (Human) | EV |
| P30530 | AXL | Tyrosine-protein kinase receptor UFO | Homo sapiens (Human) | PR |
| Q06418 | TYRO3 | Tyrosine-protein kinase receptor TYRO3 | Homo sapiens (Human) | SS |
| Q12866 | MERTK | Tyrosine-protein kinase Mer | Homo sapiens (Human) | EV |
| Q04912 | MST1R | Macrophage-stimulating protein receptor | Homo sapiens (Human) | EV |
| P34925 | RYK | Tyrosine-protein kinase RYK | Homo sapiens (Human) | PR |
| Q9QY40 | Plxnb3 | Plexin-B3 | Mus musculus (Mouse) | SS |
| P70208 | Plxna3 | Plexin-A3 | Mus musculus (Mouse) | EV SS |
| Q3UH93 | Plxnd1 | Plexin-D1 | Mus musculus (Mouse) | SS |
| P70207 | Plxna2 | Plexin-A2 | Mus musculus (Mouse) | SS |
| B2RXS4 | Plxnb2 | Plexin-B2 | Mus musculus (Mouse) | SS |
| Q8CJH3 | Plxnb1 | Plexin-B1 | Mus musculus (Mouse) | SS |
| Q9QZC2 | Plxnc1 | Plexin-C1 | Mus musculus (Mouse) | SS |
| Q80UG2 | Plxna4 | Plexin-A4 | Mus musculus (Mouse) | SS |
| P70206 | Plxna1 | Plexin-A1 | Mus musculus (Mouse) | EV SS |
| P35546 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Mus musculus (Mouse) | SS |
| Q61006 | Musk | Muscle, skeletal receptor tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q9Z138 | Ror2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Mus musculus (Mouse) | SS |
| Q9Z139 | Ror1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Mus musculus (Mouse) | PR |
| Q62371 | Ddr2 | Discoidin domain-containing receptor 2 | Mus musculus (Mouse) | SS |
| Q03146 | Ddr1 | Epithelial discoidin domain-containing receptor 1 | Mus musculus (Mouse) | SS |
| P15209 | Ntrk2 | BDNF/NT-3 growth factors receptor | Mus musculus (Mouse) | SS |
| Q3UFB7 | Ntrk1 | High affinity nerve growth factor receptor | Mus musculus (Mouse) | SS |
| Q6VNS1 | Ntrk3 | NT-3 growth factor receptor | Mus musculus (Mouse) | SS |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P35969 | Flt1 | Vascular endothelial growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P97793 | Alk | ALK tyrosine kinase receptor | Mus musculus (Mouse) | SS |
| Q9WTL4 | Insrr | Insulin receptor-related protein | Mus musculus (Mouse) | SS |
| Q60751 | Igf1r | Insulin-like growth factor 1 receptor | Mus musculus (Mouse) | SS |
| P15208 | Insr | Insulin receptor | Mus musculus (Mouse) | SS |
| P16092 | Fgfr1 | Fibroblast growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P21803 | Fgfr2 | Fibroblast growth factor receptor 2 | Mus musculus (Mouse) | SS |
| Q03142 | Fgfr4 | Fibroblast growth factor receptor 4 | Mus musculus (Mouse) | PR |
| Q60805 | Mertk | Tyrosine-protein kinase Mer | Mus musculus (Mouse) | SS |
| Q00993 | Axl | Tyrosine-protein kinase receptor UFO | Mus musculus (Mouse) | PR |
| P55144 | Tyro3 | Tyrosine-protein kinase receptor TYRO3 | Mus musculus (Mouse) | SS |
| P70424 | Erbb2 | Receptor tyrosine-protein kinase erbB-2 | Mus musculus (Mouse) | SS |
| Q61527 | Erbb4 | Receptor tyrosine-protein kinase erbB-4 | Mus musculus (Mouse) | SS |
| Q61526 | Erbb3 | Receptor tyrosine-protein kinase erbB-3 | Mus musculus (Mouse) | SS |
| Q01279 | Egfr | Epidermal growth factor receptor | Mus musculus (Mouse) | SS |
| Q01887 | Ryk | Tyrosine-protein kinase RYK | Mus musculus (Mouse) | PR |
| Q02858 | Tek | Angiopoietin-1 receptor | Mus musculus (Mouse) | SS |
| Q06806 | Tie1 | Tyrosine-protein kinase receptor Tie-1 | Mus musculus (Mouse) | SS |
| P05532 | Kit | Mast/stem cell growth factor receptor Kit | Mus musculus (Mouse) | PR |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| P09581 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Mus musculus (Mouse) | SS |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| Q00342 | Flt3 | Receptor-type tyrosine-protein kinase FLT3 | Mus musculus (Mouse) | SS |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| Q61851 | Fgfr3 | Fibroblast growth factor receptor 3 | Mus musculus (Mouse) | PR |
| Q2QLE0 | MET | Hepatocyte growth factor receptor | Sus scrofa (Pig) | PR |
| P97523 | Met | Hepatocyte growth factor receptor | Rattus norvegicus (Rat) | PR |
| Q64716 | Insrr | Insulin receptor-related protein | Rattus norvegicus (Rat) | SS |
| D3ZPX4 | Plxna3 | Plexin-A3 | Rattus norvegicus (Rat) | SS |
| D3ZLH5 | Plxnb3 | Plexin-B3 | Rattus norvegicus (Rat) | SS |
| P57097 | Mertk | Tyrosine-protein kinase Mer | Rattus norvegicus (Rat) | SS |
| Q8I7I5 | rol-3 | Protein roller-3 | Caenorhabditis elegans | PR |
| H2KZU7 | svh-2 | Tyrosine-protein kinase receptor svh-2 | Caenorhabditis elegans | SS |
| F1QVU0 | ltk | Tyrosine-protein kinase receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| F8W3R9 | alk | ALK tyrosine kinase receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6BEA0 | plxna4 | Plexin-A4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| B0S5N4 | plxna3 | Plexin A3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGLPLPLLQS | SLLLMLLLRL | SAASTNLNWQ | CPRIPYAASR | DFSVKYVVPS | FSAGGRVQAT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AAYEDSTNSA | VFVATRNHLH | VLGPDLQFIE | NLTTGPIGNP | GCQTCASCGP | GPHGPPKDTD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TLVLVMEPGL | PALVSCGSTL | QGRCFLHELE | PRGKALHLAA | PACLFSANNN | KPEACTDCVA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SPLGTRVTVV | EQGHASYFYV | ASSLDPELAA | SFSPRSVSIR | RLKSDTSGFQ | PGFPSLSVLP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KYLASYLIKY | VYSFHSGDFV | YFLTVQPISV | TSPPSALHTR | LVRLNAVEPE | IGDYRELVLD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| CHFAPKRRRR | GAPEGTQPYP | VLQAAHSAPV | DAKLAVELSI | SEGQEVLFGV | FVTVKDGGSG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MGPNSVVCAF | PIYHLNILIE | EGVEYCCHSS | NSSSLLSRGL | DFFQTPSFCP | NPPGGEASGP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SSRCHYFPLM | VHASFTRVDL | FNGLLGSVKV | TALHVTRLGN | VTVAHMGTVD | GRVLQVEIAR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SLNYLLYVSN | FSLGSSGQPV | HRDVSRLGND | LLFASGDQVF | KVPIQGPGCR | HFLTCWRCLR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AQRFMGCGWC | GDRCDRQKEC | PGSWQQDHCP | PEISEFYPHS | GPLRGTTRLT | LCGSNFYLRP |
| 610 | 620 | 630 | 640 | 650 | 660 |
| DDVVPEGTHQ | ITVGQSPCRL | LPKDSSSPRP | GSLKEFIQEL | ECELEPLVTQ | AVGTTNISLV |
| 670 | 680 | 690 | 700 | 710 | 720 |
| ITNMPAGKHF | RVEGISVQEG | FSFVEPVLTS | IKPDFGPRAG | GTYLTLEGQS | LSVGTSRAVL |
| 730 | 740 | 750 | 760 | 770 | 780 |
| VNGTQCRLEQ | VNEEQILCVT | PPGAGTARVP | LHLQIGGAEV | PGSWTFHYKE | DPIVLDISPK |
| 790 | 800 | 810 | 820 | 830 | 840 |
| CGYSGSHIMI | HGQHLTSAWH | FTLSFHDGQS | TVESRCAGQF | VEQQQRRCRL | PEYVVRNPQG |
| 850 | 860 | 870 | 880 | 890 | 900 |
| WATGNLSVWG | DGAAGFTLPG | FRFLPPPSPL | RAGLVELKPE | EHSVKVEYVG | LGAVADCVTV |
| 910 | 920 | 930 | 940 | 950 | 960 |
| NMTVGGEVCQ | HELRGDVVIC | PLPPSLQLGK | DGVPLQVCVD | GGCHILSQVV | RSSPGRASQR |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| ILLIALLVLI | LLVAVLAVAL | IFNSRRRKKQ | LGAHSLSPTT | LSDINDTASG | APNHEESSES |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| RDGTSVPLLR | TESIRLQDLD | RMLLAEVKDV | LIPHEQVVIH | TDQVIGKGHF | GVVYHGEYTD |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| GAQNQTHCAI | KSLSRITEVQ | EVEAFLREGL | LMRGLHHPNI | LALIGIMLPP | EGLPRVLLPY |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| MRHGDLLHFI | RSPQRNPTVK | DLVSFGLQVA | CGMEYLAEQK | FVHRDLAARN | CMLDESFTVK |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| VADFGLARGV | LDKEYYSVRQ | HRHARLPVKW | MALESLQTYR | FTTKSDVWSF | GVLLWELLTR |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| GAPPYPHIDP | FDLSHFLAQG | RRLPQPEYCP | DSLYHVMLRC | WEADPAARPT | FRALVLEVKQ |
| 1330 | 1340 | 1350 | 1360 | 1370 | |
| VVASLLGDHY | VQLTAAYVNV | GPRAVDDGSV | PPEQVQPSPQ | HCRSTSKPRP | LSEPPLPT |