Q61084
Gene name |
Map3k3 (Mekk3) |
Protein name |
Mitogen-activated protein kinase kinase kinase 3 |
Names |
MAPK/ERK kinase kinase 3, MEK kinase 3, MEKK 3 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:26406 |
EC number |
2.7.11.25: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
506-532 (Activation loop from InterPro)
Target domain |
362-622 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
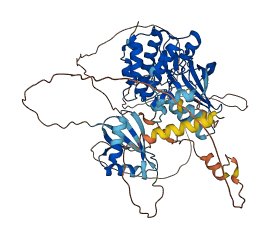
Autoinhibited structure

Activated structure

1 structures for Q61084
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q61084-F1 | Predicted | AlphaFoldDB |
25 variants for Q61084
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3402727653 | 2 | D>V | No | EVA | |
| rs3389210122 | 51 | H>R | No | EVA | |
| rs3389209736 | 102 | K>N | No | EVA | |
| rs3389221940 | 110 | S>N | No | EVA | |
| rs3389181211 | 114 | K>R | No | EVA | |
| rs3389224508 | 115 | S>G | No | EVA | |
| rs3389206596 | 132 | P>L | No | EVA | |
| rs3389198375 | 146 | Q>H | No | EVA | |
| rs3389146788 | 159 | E>G | No | EVA | |
| rs3389214446 | 250 | S>R | No | EVA | |
| rs3402634617 | 307 | T>A | No | EVA | |
| rs3402845749 | 311 | S>P | No | EVA | |
| rs3389221915 | 346 | V>M | No | EVA | |
| rs3389213143 | 403 | S>N | No | EVA | |
| rs3389204319 | 419 | L>M | No | EVA | |
| rs230741806 | 451 | V>I | No | EVA | |
| rs3402754934 | 520 | S>P | No | EVA | |
| rs3402634640 | 521 | G>V | No | EVA | |
| rs260838428 | 524 | I>M | No | EVA | |
| rs3389210139 | 532 | Y>* | No | EVA | |
| rs3389206619 | 535 | S>R | No | EVA | |
| rs3389219000 | 542 | E>D | No | EVA | |
| rs3389206560 | 592 | I>N | No | EVA | |
| rs3389185492 | 601 | R>K | No | EVA | |
| rs3389209760 | 619 | H>Q | No | EVA |
No associated diseases with Q61084
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.25 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| MAP kinase kinase kinase activity | Catalysis of the phosphorylation and activation of a MAP kinase kinase; each MAP kinase kinase can be phosphorylated by any of several MAP kinase kinase kinases. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| blood vessel development | The process whose specific outcome is the progression of a blood vessel over time, from its formation to the mature structure. The blood vessel is the vasculature carrying blood. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of cellular senescence | Any process that stops, prevents or reduces the frequency, rate or extent of cellular senescence. |
| positive regulation of cell migration involved in sprouting angiogenesis | Any process that increases the frequency, rate or extent of cell migration involved in sprouting angiogenesis. Cell migration involved in sprouting angiogenesis is the orderly movement of endothelial cells into the extracellular matrix in order to form new blood vessels contributing to the process of sprouting angiogenesis. |
| positive regulation of cell proliferation in bone marrow | A process that activates or increases the frequency, rate or extent of cell proliferation in the bone marrow. |
| positive regulation of I-kappaB kinase/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| positive regulation of p38MAPK cascade | Any process that activates or increases the frequency, rate or extent of p38MAPK cascade. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9Y2U5 | MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | Homo sapiens (Human) | PR |
| Q99759 | MAP3K3 | Mitogen-activated protein kinase kinase kinase 3 | Homo sapiens (Human) | PR |
| Q9ES70 | Nek6 | Serine/threonine-protein kinase Nek6 | Mus musculus (Mouse) | SS |
| Q61083 | Map3k2 | Mitogen-activated protein kinase kinase kinase 2 | Mus musculus (Mouse) | PR |
| O22042 | ANP3 | Mitogen-activated protein kinase kinase kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDEQEALDSI | MKDLVALQMS | RRTRLSGYET | MKNKDTGHPN | RQSDVRIKFE | HNGERRIIAF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SRPVRYEDVE | HKVTTVFGQP | LDLHYMNNEL | SILLKNQDDL | DKAIDILDRS | SSMKSLRILL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LSQDRNHTSS | SPHSGVSRQV | RIKPSQSAGD | INTIYQAPEP | RSRHLSVSSQ | NPGRSSPPPG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YVPERQQHIA | RQGSYTSINS | EGEFIPETSE | QCMLDPLSSA | ENSLSGSCQS | LDRSADSPSF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RKSQMSRARS | FPDNRKECSD | RETQLYDKGV | KGGTYPRRYH | VSVHHKDYND | GRRTFPRIRR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HQGNLFTLVP | SSRSLSTNGE | NMGVAVQYLD | PRGRLRSADS | ENALTVQERN | VPTKSPSAPI |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NWRRGKLLGQ | GAFGRVYLCY | DVDTGRELAS | KQVQFDPDSP | ETSKEVSALE | CEIQLLKNLQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| HERIVQYYGC | LRDRAEKILT | IFMEYMPGGS | VKDQLKAYGA | LTESVTRKYT | RQILEGMSYL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| HSNMIVHRDI | KGANILRDSA | GNVKLGDFGA | SKRLQTICMS | GTGIRSVTGT | PYWMSPEVIS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GEGYGRKADV | WSLGCTVVEM | LTEKPPWAEY | EAMAAIFKIA | TQPTNPQLPS | HISEHGRDFL |
| 610 | 620 | ||||
| RRIFVEARQR | PSAEELLTHH | FAQLVY |