Q60992
Gene name |
Vav2 |
Protein name |
Guanine nucleotide exchange factor VAV2 |
Names |
VAV-2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:22325 |
EC number |
|
Protein Class |
|
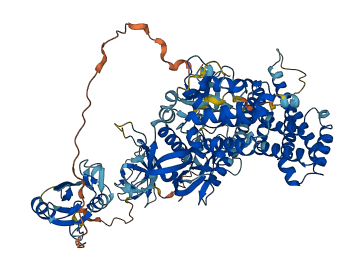
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
193-371 (DH domain) |
Relief mechanism |
Partner binding, PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Aghazadeh B et al. (2000) "Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation", Cell, 102, 625-33
- Barreira M et al. (2014) "The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins", Science signaling, 7, ra35
- Yu B et al. (2010) "Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1", Cell, 140, 246-56
- Rapley J et al. (2008) "Crucial structural role for the PH and C1 domains of the Vav1 exchange factor", EMBO reports, 9, 655-61
Autoinhibited structure

Activated structure

1 structures for Q60992
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q60992-F1 | Predicted | AlphaFoldDB |
44 variants for Q60992
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388535652 | 75 | I>S | No | EVA | |
| rs3388540851 | 77 | T>I | No | EVA | |
| rs3388538827 | 81 | V>F | No | EVA | |
| rs3388542112 | 138 | D>V | No | EVA | |
| rs3388540550 | 141 | V>I | No | EVA | |
| rs3388538922 | 157 | D>N | No | EVA | |
| rs3388537903 | 169 | D>G | No | EVA | |
| rs3388537797 | 179 | E>* | No | EVA | |
| rs3388537916 | 181 | Q>K | No | EVA | |
| rs3388540552 | 189 | T>I | No | EVA | |
| rs3388538232 | 200 | E>V | No | EVA | |
| rs3388532265 | 235 | V>I | No | EVA | |
| rs3388532211 | 246 | H>Y | No | EVA | |
| rs3388532205 | 255 | V>M | No | EVA | |
| rs3388542086 | 265 | A>D | No | EVA | |
| rs3388537835 | 283 | S>N | No | EVA | |
| rs3388540862 | 287 | H>Y | No | EVA | |
| rs261140679 | 295 | L>V | No | EVA | |
| rs3388532221 | 350 | E>K | No | EVA | |
| rs3388538786 | 404 | D>E | No | EVA | |
| rs229567277 | 449 | F>L | No | EVA | |
| rs3391247590 | 451 | K>T | No | EVA | |
| rs3391377053 | 454 | D>N | No | EVA | |
| rs3388538193 | 508 | K>N | No | EVA | |
| rs3388537807 | 520 | T>I | No | EVA | |
| rs3388537949 | 524 | T>S | No | EVA | |
| rs3388538602 | 552 | H>D | No | EVA | |
| rs3388538602 | 552 | H>Y | No | EVA | |
| rs3388538872 | 558 | V>A | No | EVA | |
| rs3388537909 | 559 | I>F | No | EVA | |
| rs3388537888 | 593 | P>L | No | EVA | |
| rs3388538223 | 596 | K>R | No | EVA | |
| rs3388540548 | 626 | T>I | No | EVA | |
| rs3388540849 | 628 | K>R | No | EVA | |
| rs3388537814 | 678 | K>N | No | EVA | |
| rs3388537866 | 712 | V>SDG* | No | EVA | |
| rs3388538207 | 740 | H>Y | No | EVA | |
| rs3388532262 | 745 | S>R | No | EVA | |
| rs864293105 | 763 | T>S | No | EVA | |
| rs3388538824 | 773 | A>T | No | EVA | |
| rs3391310758 | 788 | L>P | No | EVA | |
| rs3388538239 | 807 | V>G | No | EVA | |
| rs3388538628 | 812 | V>M | No | EVA | |
| rs3388541181 | 824 | E>G | No | EVA |
No associated diseases with Q60992
13 regional properties for Q60992
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Dbl homology (DH) domain | 193 - 371 | IPR000219 |
| domain | SH2 domain | 661 - 757 | IPR000980 |
| conserved_site | Guanine-nucleotide dissociation stimulator, CDC24, conserved site | 320 - 345 | IPR001331 |
| domain | SH3 domain | 576 - 642 | IPR001452-1 |
| domain | SH3 domain | 806 - 867 | IPR001452-2 |
| domain | Calponin homology domain | 1 - 120 | IPR001715 |
| domain | Pleckstrin homology domain | 400 - 504 | IPR001849 |
| domain | Protein kinase C-like, phorbol ester/diacylglycerol-binding domain | 513 - 562 | IPR002219 |
| domain | Calmodulin-regulated spectrin-associated protein-like, Calponin-homology domain | 19 - 103 | IPR022613 |
| domain | VAV2 protein, second SH3 domain | 809 - 866 | IPR035732 |
| domain | VAV2 protein, first SH3 domain | 580 - 639 | IPR035733 |
| domain | VAV2, SH2 domain | 657 - 759 | IPR035880 |
| domain | Vav, PH domain | 384 - 506 | IPR037832 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| epidermal growth factor receptor binding | Binding to an epidermal growth factor receptor. |
| guanyl-nucleotide exchange factor activity | Stimulates the exchange of GDP to GTP on a signaling GTPase, changing its conformation to its active form. Guanine nucleotide exchange factors (GEFs) act by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP), which is more abundant in the cell under normal cellular physiological conditions. |
| metal ion binding | Binding to a metal ion. |
| phosphotyrosine residue binding | Binding to a phosphorylated tyrosine residue within a protein. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. |
| cell projection assembly | Formation of a prolongation or process extending from a cell, e.g. a flagellum or axon. |
| lamellipodium assembly | Formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| regulation of cell size | Any process that modulates the size of a cell. |
| small GTPase-mediated signal transduction | The series of molecular signals in which a small monomeric GTPase relays a signal. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9NHV9 | Vav | Protein vav | Drosophila melanogaster (Fruit fly) | SS |
| P15498 | VAV1 | Proto-oncogene vav | Homo sapiens (Human) | EV SS |
| Q9UKW4 | VAV3 | Guanine nucleotide exchange factor VAV3 | Homo sapiens (Human) | SS |
| P52735 | VAV2 | Guanine nucleotide exchange factor VAV2 | Homo sapiens (Human) | SS |
| P27870 | Vav1 | Proto-oncogene vav | Mus musculus (Mouse) | EV |
| Q9R0C8 | Vav3 | Guanine nucleotide exchange factor VAV3 | Mus musculus (Mouse) | SS |
| P54100 | Vav1 | Proto-oncogene vav | Rattus norvegicus (Rat) | SS |
| Q45FX5 | vav-1 | Protein vav-1 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEQWRQCGRW | LIDCKVLPPN | HRVVWPSAVV | FDLAQALRDG | VLLCQLLHNL | SPGSIDLKDI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NFRPQMSQFL | CLKNIRTFLK | VCHDKFGLRN | SELFDPFDLF | DVRDFGKVIS | AVSRLSLHSI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AQSKGIRPFP | SEETAENDDD | VYRSLEELAD | EHDLGEDIYD | CVPCEDEGDD | IYEDIIKVEV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QQPMKMGMTE | DDKRSCCLLE | IQETEAKYYR | TLEDIEKNYM | GPLRLVLSPA | DMAAVFINLE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DLIKVHHSFL | RAIDVSMMAG | GSTLAKVFLE | FKERLLIYGE | YCSHMEHAQS | TLNQLLASRE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DFRQKVEECT | LRVQDGKFKL | QDLLVVPMQR | VLKYHLLLKE | LLSHSADRPE | RQQLKEALEA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MQDLAMYINE | VKRDKETLKK | ISEFQCSIEN | LQVKLEEFGR | PKIDGELKVR | SIVNHTKQDR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YLFLFDKVVI | VCKRKGYSYE | LKEVIELLFH | KMTDDPMHNK | DIKKWSYGFY | LIHLQGKQGF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QFFCKTEDMK | RKWMEQFEMA | MSNIKPDKAN | ANHHSFQMYT | FDKTTNCKAC | KMFLRGTFYQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GYLCTRCGVG | AHKECLEVIP | PCKMSSPADV | DAPGAGPGPK | MVAVQNYHGN | PAPPGKPVLT |
| 610 | 620 | 630 | 640 | 650 | 660 |
| FQTGDVIELL | RGDPDSPWWE | GRLVQTRKSG | YFPSSSVKPC | PVDGRPPTGR | PPSREIDYTA |
| 670 | 680 | 690 | 700 | 710 | 720 |
| YPWFAGNMER | QQTDNLLKSH | ASGTYLIRER | PAEAERFAIS | IKFNDEVKHI | KVVEKDSWIH |
| 730 | 740 | 750 | 760 | 770 | 780 |
| ITEAKKFESL | LELVEYYQCH | SLKESFKQLD | TTLKFPYKSR | ERTTSRASSR | SPASCASYNF |
| 790 | 800 | 810 | 820 | 830 | 840 |
| SFLSPQGLSF | APQAPSAPFW | SVFTPRVIGT | AVARYNFAAR | DMRELSLREG | DVVKIYSRIG |
| 850 | 860 | ||||
| GDQGWWKGET | NGRIGWFPST | YVEEEGVQ |