Q60823
Gene name |
Akt2 |
Protein name |
RAC-beta serine/threonine-protein kinase |
Names |
Protein kinase Akt-2, Protein kinase B beta, PKB beta, RAC-PK-beta |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:11652 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with P31749)
The protein kinase Akt is one of the primary effectors of growth factor signaling in the cell. Akt responds specifically to the lipid second messengers phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3] and phosphatidylinositol-3,4-bisphosphate [PI(3,4)P2] via its autoinhibitory domain (PH domain). Recruitment of Akt to PI(3,4,5)P3 in the plasma membrane promotes its phosphorylation by phosphoinositide-dependent kinase 1 (PDK1) in its activation loop (T308). Phosphorylation of S473 within AGC kinase C-terminal domain activates Akt through the formation of an electrostatic interaction with a conserved basic residue (R144) in the PH-kinase domain linker, thereby relieving PH domain- mediated autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
292-315 (Activation loop from InterPro)
Target domain |
156-478 (Catalytic domain of the Serine/Threonine Kinase, Protein Kinase B beta, also called Akt2) |
Relief mechanism |
|
Assay |
|
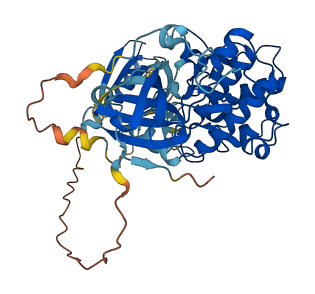
Autoinhibited structure

Activated structure

1 structures for Q60823
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q60823-F1 | Predicted | AlphaFoldDB |
25 variants for Q60823
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388895066 | 35 | F>I | No | EVA | |
| rs3388851305 | 55 | F>L | No | EVA | |
| rs3388872404 | 77 | C>W | No | EVA | |
| rs233720581 | 94 | D>E | No | EVA | |
| rs237667590 | 101 | R>Q | No | EVA | |
| rs3388875040 | 169 | V>D | No | EVA | |
| rs3411787349 | 181 | K>M | No | EVA | |
| rs3388892329 | 197 | T>R | No | EVA | |
| rs3388893870 | 202 | R>L | No | EVA | |
| rs3388884849 | 212 | L>I | No | EVA | |
| rs3388872396 | 229 | M>T | No | EVA | |
| rs3388875025 | 241 | L>F | No | EVA | |
| rs3388884764 | 242 | S>F | No | EVA | |
| rs3397688368 | 265 | Y>* | No | EVA | |
| rs3397688332 | 267 | H>N | No | EVA | |
| rs3388891563 | 293 | D>E | No | EVA | |
| rs3388865782 | 294 | F>L | No | EVA | |
| rs3388886367 | 308 | K>R | No | EVA | |
| rs3388897280 | 309 | T>I | No | EVA | |
| rs3388851343 | 337 | G>D | No | EVA | |
| rs3388872375 | 373 | L>F | No | EVA | |
| rs3388887481 | 377 | A>V | No | EVA | |
| rs3388897228 | 421 | K>Q | No | EVA | |
| rs3388892743 | 472 | Q>R | No | EVA | |
| rs3388865867 | 480 | R>Q | No | EVA |
No associated diseases with Q60823
8 regional properties for Q60823
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 152 - 409 | IPR000719 |
| domain | AGC-kinase, C-terminal | 410 - 481 | IPR000961 |
| domain | Pleckstrin homology domain | 5 - 110 | IPR001849 |
| active_site | Serine/threonine-protein kinase, active site | 271 - 283 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 158 - 191 | IPR017441 |
| domain | Protein kinase, C-terminal | 430 - 475 | IPR017892 |
| domain | Protein Kinase B beta, catalytic domain | 156 - 478 | IPR034677 |
| domain | Protein Kinase B, pleckstrin homology domain | 4 - 111 | IPR039026 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| insulin-responsive compartment | A small membrane-bounded vesicle that releases its contents by exocytosis in response to insulin stimulation; the contents are enriched in GLUT4, IRAP and VAMP2. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
| sarcoplasmic reticulum | A fine reticular network of membrane-limited elements that pervades the sarcoplasm of a muscle cell; continuous over large portions of the cell and with the nuclear envelope; that part of the endoplasmic reticulum specialized for calcium release, uptake and storage. |
| vesicle | Any small, fluid-filled, spherical organelle enclosed by membrane. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein kinase C binding | Binding to protein kinase C. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
40 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of GTPase activity | Any process that initiates the activity of an inactive GTPase through the replacement of GDP by GTP. |
| carbohydrate transport | The directed movement of carbohydrate into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. Carbohydrates are a group of organic compounds based of the general formula Cx(H2O)y. |
| cellular response to high light intensity | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a high light intensity stimulus. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| cellular response to organic cyclic compound | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic cyclic compound stimulus. |
| glucose metabolic process | The chemical reactions and pathways involving glucose, the aldohexose gluco-hexose. D-glucose is dextrorotatory and is sometimes known as dextrose; it is an important source of energy for living organisms and is found free as well as combined in homo- and hetero-oligosaccharides and polysaccharides. |
| glycogen biosynthetic process | The chemical reactions and pathways resulting in the formation of glycogen, a polydisperse, highly branched glucan composed of chains of D-glucose residues. |
| insulin receptor signaling pathway | The series of molecular signals generated as a consequence of the insulin receptor binding to insulin. |
| intracellular protein transmembrane transport | The directed movement of proteins in a cell, from one side of a membrane to another by means of some agent such as a transporter or pore. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of a cysteine-type endopeptidase activity involved in the apoptotic process. |
| negative regulation of long-chain fatty acid import across plasma membrane | Any process that decreases the rate, frequency or extent of plasma membrane long-chain fatty acid transport. Plasma membrane long-chain fatty acid transport is the directed movement of long-chain fatty acids across the plasma membrane. |
| negative regulation of RNA splicing | Any process that stops, prevents, or reduces the frequency, rate or extent of RNA splicing. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| peripheral nervous system myelin maintenance | The process in which the structure and material content of mature peripheral nervous system myelin is kept in a functional state. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell motility | Any process that activates or increases the frequency, rate or extent of cell motility. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of fatty acid beta-oxidation | Any process that activates or increases the frequency, rate or extent of fatty acid beta-oxidation. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of glucose import | Any process that activates or increases the frequency, rate or extent of the import of the hexose monosaccharide glucose into a cell or organelle. |
| positive regulation of glucose metabolic process | Any process that increases the rate, frequency or extent of glucose metabolism. Glucose metabolic processes are the chemical reactions and pathways involving glucose, the aldohexose gluco-hexose. |
| positive regulation of glycogen biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of glycogen. |
| positive regulation of mitochondrial membrane potential | Any process that activates or increases the frequency, rate or extent of establishment or extent of a mitochondrial membrane potential, the electric potential existing across any mitochondrial membrane arising from charges in the membrane itself and from the charges present in the media on either side of the membrane. |
| positive regulation of nitric oxide biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nitric oxide. |
| positive regulation of peptidyl-serine phosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| positive regulation of positive chemotaxis | Any process that activates or increases the frequency, rate or extent of the directed movement of a motile cell or organism towards a higher concentration in a concentration gradient of a specific chemical. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| positive regulation of protein targeting to membrane | Any process that increases the frequency, rate or extent of the process of directing proteins towards a membrane, usually using signals contained within the protein. |
| positive regulation of signal transduction | Any process that activates or increases the frequency, rate or extent of signal transduction. |
| positive regulation of sodium ion transport | Any process that increases the frequency, rate or extent of the directed movement of sodium ions (Na+) into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of vesicle fusion | Any process that activates or increases the frequency, rate or extent of vesicle fusion. |
| protein kinase B signaling | A series of reactions, mediated by the intracellular serine/threonine kinase protein kinase B (also called AKT), which occurs as a result of a single trigger reaction or compound. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of translation | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| response to insulin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| retinal rod cell apoptotic process | Any apoptotic process in a retinal rod cell, one of the two photoreceptor cell types of the vertebrate retina. |
29 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q01314 | AKT1 | RAC-alpha serine/threonine-protein kinase | Bos taurus (Bovine) | SS |
| Q8INB9 | Akt | RAC serine/threonine-protein kinase | Drosophila melanogaster (Fruit fly) | SS |
| Q9Y243 | AKT3 | RAC-gamma serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| P31751 | AKT2 | RAC-beta serine/threonine-protein kinase | Homo sapiens (Human) | EV SS |
| P31750 | Akt1 | RAC-alpha serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q62074 | Prkci | Protein kinase C iota type | Mus musculus (Mouse) | SS |
| Q9Z2A0 | Pdpk1 | 3-phosphoinositide-dependent protein kinase 1 | Mus musculus (Mouse) | SS |
| Q02956 | Prkcz | Protein kinase C zeta type | Mus musculus (Mouse) | SS |
| P70268 | Pkn1 | Serine/threonine-protein kinase N1 | Mus musculus (Mouse) | SS |
| Q8K045 | Pkn3 | Serine/threonine-protein kinase N3 | Mus musculus (Mouse) | SS |
| Q8BWW9 | Pkn2 | Serine/threonine-protein kinase N2 | Mus musculus (Mouse) | SS |
| P23298 | Prkch | Protein kinase C eta type | Mus musculus (Mouse) | PR |
| P16054 | Prkce | Protein kinase C epsilon type | Mus musculus (Mouse) | PR |
| Q02111 | Prkcq | Protein kinase C theta type | Mus musculus (Mouse) | PR |
| P28867 | Prkcd | Protein kinase C delta type | Mus musculus (Mouse) | PR |
| P20444 | Prkca | Protein kinase C alpha type | Mus musculus (Mouse) | SS |
| P63318 | Prkcg | Protein kinase C gamma type | Mus musculus (Mouse) | SS |
| P68404 | Prkcb | Protein kinase C beta type | Mus musculus (Mouse) | SS |
| Q9WUA6 | Akt3 | RAC-gamma serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q9ERE3 | Sgk3 | Serine/threonine-protein kinase Sgk3 | Mus musculus (Mouse) | PR |
| Q8QZV4 | Stk32c | Serine/threonine-protein kinase 32C | Mus musculus (Mouse) | PR |
| Q91VJ4 | Stk38 | Serine/threonine-protein kinase 38 | Mus musculus (Mouse) | SS |
| Q7TSE6 | Stk38l | Serine/threonine-protein kinase 38-like | Mus musculus (Mouse) | SS |
| P47196 | Akt1 | RAC-alpha serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| Q63484 | Akt3 | RAC-gamma serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| P47197 | Akt2 | RAC-beta serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| Q17941 | akt-1 | Serine/threonine-protein kinase akt-1 | Caenorhabditis elegans | PR |
| Q9XTG7 | akt-2 | Serine/threonine-protein kinase akt-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNEVSVIKEG | WLHKRGEYIK | TWRPRYFLLK | SDGSFIGYKE | RPEAPDQTLP | PLNNFSVAEC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QLMKTERPRP | NTFVIRCLQW | TTVIERTFHV | DSPDEREEWM | RAIQMVANSL | KQRGPGEDAM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DYKCGSPSDS | STSEMMEVAV | NKARAKVTMN | DFDYLKLLGK | GTFGKVILVR | EKATGRYYAM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KILRKEVIIA | KDEVAHTVTE | SRVLQNTRHP | FLTALKYAFQ | THDRLCFVME | YANGGELFFH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LSRERVFTED | RARFYGAEIV | SALEYLHSRD | VVYRDIKLEN | LMLDKDGHIK | ITDFGLCKEG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ISDGATMKTF | CGTPEYLAPE | VLEDNDYGRA | VDWWGLGVVM | YEMMCGRLPF | YNQDHERLFE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LILMEEIRFP | RTLGPEAKSL | LAGLLKKDPK | QRLGGGPSDA | KEVMEHRFFL | SINWQDVVQK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KLLPPFKPQV | TSEVDTRYFD | DEFTAQSITI | TPPDRYDSLD | PLELDQRTHF | PQFSYSASIR |
| E |