Q60751
Gene name |
Igf1r |
Protein name |
Insulin-like growth factor 1 receptor |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:16001 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
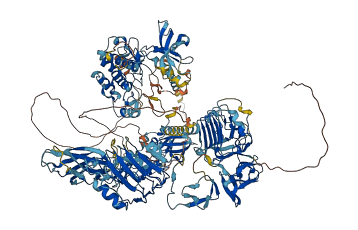
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1000-1276 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
1154-1179 (Activation loop from InterPro)
Target domain |
1000-1276 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Craddock BP et al. (2007) "Autoinhibition of the insulin-like growth factor I receptor by the juxtamembrane region", FEBS letters, 581, 3235-40
- Uchikawa E et al. (2019) "Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex", eLife, 8,
- Nielsen J et al. (2022) "Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling", Journal of molecular biology, 434, 167458
- Chen YS et al. (2021) "Insertion of a synthetic switch into insulin provides metabolite-dependent regulation of hormone-receptor activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
Autoinhibited structure

Activated structure

3 structures for Q60751
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6PYH | EM | 430 A | A/D | 31-1292 | PDB |
| 8EYR | EM | 400 A | A/B | 31-1292 | PDB |
| AF-Q60751-F1 | Predicted | AlphaFoldDB |
No variants for Q60751
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q60751 | |||||
No associated diseases with Q60751
4 regional properties for Q60751
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Cation-transporting P-type ATPase, N-terminal | 99 - 176 | IPR004014 |
| domain | Cation-transporting P-type ATPase, C-terminal | 830 - 999 | IPR006068 |
| ptm | P-type ATPase, phosphorylation site | 449 - 455 | IPR018303 |
| domain | P-type ATPase, haloacid dehalogenase domain | 429 - 798 | IPR044492 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
16 GO annotations of cellular component
| Name | Definition |
|---|---|
| alphav-beta3 integrin-IGF-1-IGF1R complex | A protein complex that consists of an alphav-beta3 integrin complex bound to insulin-like growth factor-1 (IGF-1) and type I insulin-like growth factor receptor (IGF1R). IGF1R is a heterotetramer that consists of two alpha-subunits and two beta-subunits. |
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| caveola | A membrane raft that forms small pit, depression, or invagination that communicates with the outside of a cell and extends inward, indenting the cytoplasm and the cell membrane. Examples include flask-shaped invaginations of the plasma membrane in adipocytes associated with caveolin proteins, and minute pits or incuppings of the cell membrane formed during pinocytosis. Caveolae may be pinched off to form free vesicles within the cytoplasm. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| insulin receptor complex | A disulfide-bonded, heterotetrameric receptor complex. The alpha chains are entirely extracellular, while each beta chain has one transmembrane domain. The ligand binds to the alpha subunit extracellular domain and the kinase is associated with the beta subunit intracellular domain. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein kinase complex | A protein complex which is capable of protein kinase activity. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| T-tubule | Invagination of the plasma membrane of a muscle cell that extends inward from the cell surface around each myofibril. The ends of T-tubules make contact with the sarcoplasmic reticulum membrane. |
17 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| G-protein alpha-subunit binding | Binding to a G-protein alpha subunit. The alpha subunit binds a guanine nucleotide. |
| identical protein binding | Binding to an identical protein or proteins. |
| insulin binding | Binding to insulin, a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| insulin receptor binding | Binding to an insulin receptor. |
| insulin receptor substrate binding | Binding to an insulin receptor substrate (IRS) protein, an adaptor protein that bind to the transphosphorylated insulin and insulin-like growth factor receptors, are themselves phosphorylated and in turn recruit SH2 domain-containing signaling molecules to form a productive signaling complex. |
| insulin-activated receptor activity | Combining with insulin receptor ligand and transmitting the signal across the plasma membrane to initiate a change in cell activity. |
| insulin-like growth factor binding | Binding to an insulin-like growth factor, any member of a group of polypeptides that are structurally homologous to insulin and share many of its biological activities, but are immunologically distinct from it. |
| insulin-like growth factor I binding | Binding to insulin-like growth factor I. |
| insulin-like growth factor-activated receptor activity | Combining with insulin-like growth factor receptor ligand and transmitting the signal across the plasma membrane to initiate a change in cell activity. |
| phosphatidylinositol 3-kinase binding | Binding to a phosphatidylinositol 3-kinase, any enzyme that catalyzes the addition of a phosphate group to an inositol lipid at the 3' position of the inositol ring. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein transporter activity | Directly binding to a specific protein and delivering it to a specific cellular location. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| structural molecule activity | The action of a molecule that contributes to the structural integrity of a complex or its assembly within or outside a cell. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
58 GO annotations of biological process
| Name | Definition |
|---|---|
| adrenal gland development | The process whose specific outcome is the progression of the adrenal gland over time, from its formation to the mature structure. This gland can either be a discrete structure located bilaterally above each kidney, or a cluster of cells in the head kidney that perform the functions of the adrenal gland. In either case, this organ consists of two cells types, aminergic chromaffin cells and steroidogenic cortical cells. |
| amyloid-beta clearance | The process in which amyloid-beta is removed from extracellular brain regions by mechanisms involving cell surface receptors. |
| animal organ morphogenesis | Morphogenesis of an animal organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process in which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions. |
| axonogenesis | De novo generation of a long process of a neuron, including the terminal branched region. Refers to the morphogenesis or creation of shape or form of the developing axon, which carries efferent (outgoing) action potentials from the cell body towards target cells. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| cellular response to amyloid-beta | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a amyloid-beta stimulus. |
| cellular response to glucose stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus. |
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| cellular response to insulin-like growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin-like growth factor stimulus. |
| cellular response to transforming growth factor beta stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a transforming growth factor beta stimulus. |
| cerebellum development | The process whose specific outcome is the progression of the cerebellum over time, from its formation to the mature structure. The cerebellum is the portion of the brain in the back of the head between the cerebrum and the pons. In mice, the cerebellum controls balance for walking and standing, modulates the force and range of movement and is involved in the learning of motor skills. |
| dendritic spine maintenance | The organization process that preserves a dendritic spine in a stable functional or structural state. A dendritic spine is a specialized protrusion from a neuronal dendrite and is involved in synaptic transmission. |
| epidermis development | The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of an animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species. |
| establishment of cell polarity | The specification and formation of anisotropic intracellular organization or cell growth patterns. |
| estrous cycle | A type of ovulation cycle, which occurs in most mammalian therian females, where the endometrium is resorbed if pregnancy does not occur. |
| exocrine pancreas development | The process whose specific outcome is the progression of the exocrine pancreas over time, from its formation to the mature structure. The exocrine pancreas produces and store zymogens of digestive enzymes, such as chymotrypsinogen and trypsinogen in the acinar cells. |
| glucose homeostasis | Any process involved in the maintenance of an internal steady state of glucose within an organism or cell. |
| immune response | Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat. |
| insulin receptor signaling pathway | The series of molecular signals generated as a consequence of the insulin receptor binding to insulin. |
| insulin-like growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to an insulin-like growth factor receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| male sex determination | The specification of male sex of an individual organism. |
| mammary gland development | The process whose specific outcome is the progression of the mammary gland over time, from its formation to the mature structure. The mammary gland is a large compound sebaceous gland that in female mammals is modified to secrete milk. Its development starts with the formation of the mammary line and ends as the mature gland cycles between nursing and weaning stages. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of cholangiocyte apoptotic process | Any process that stops, prevents or reduces the frequency, rate or extent of cholangiocyte apoptotic process. |
| negative regulation of DNA-binding transcription factor activity | Any process that stops, prevents, or reduces the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| negative regulation of hepatocyte apoptotic process | Any process that stops, prevents or reduces the frequency, rate or extent of hepatocyte apoptotic process. |
| negative regulation of MAPK cascade | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction mediated by the MAPKKK cascade. |
| negative regulation of muscle cell apoptotic process | Any process that decreases the rate or frequency of muscle cell apoptotic process, a form of programmed cell death induced by external or internal signals that trigger the activity of proteolytic caspases whose actions dismantle a muscle cell and result in its death. |
| negative regulation of protein kinase B signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| neuron projection development | The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| peptidyl-tyrosine autophosphorylation | The phosphorylation by a protein of one or more of its own tyrosine amino acid residues, or a tyrosine residue on an identical protein. |
| phosphatidylinositol-mediated signaling | The series of molecular signals in which a cell uses a phosphatidylinositol-mediated signaling to convert a signal into a response. Phosphatidylinositols include phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| positive regulation of axon regeneration | Any process that activates, maintains or increases the rate of axon regeneration. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of cold-induced thermogenesis | Any process that activates or increases the frequency, rate or extent of cold-induced thermogenesis. |
| positive regulation of cytokinesis | Any process that activates or increases the frequency, rate or extent of the division of the cytoplasm of a cell, and its separation into two daughter cells. |
| positive regulation of developmental growth | Any process that activates, maintains or increases the rate of developmental growth. |
| positive regulation of DNA metabolic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving DNA. |
| positive regulation of kinase activity | Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| positive regulation of meiotic cell cycle | Any process that activates or increases the frequency, rate or extent of progression through the meiotic cell cycle. |
| positive regulation of mitotic nuclear division | Any process that activates or increases the frequency, rate or extent of mitosis. |
| positive regulation of osteoblast proliferation | Any process that activates or increases the rate or extent of osteoblast proliferation. |
| positive regulation of phosphatidylinositol 3-kinase signaling | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the phosphatidylinositol 3-kinase cascade. |
| positive regulation of protein kinase B signaling | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| positive regulation of protein-containing complex disassembly | Any process that activates or increases the frequency, rate or extent of protein complex disassembly, the disaggregation of a protein complex into its constituent components. |
| positive regulation of smooth muscle cell proliferation | Any process that activates or increases the rate or extent of smooth muscle cell proliferation. |
| positive regulation of steroid hormone biosynthetic process | Any process that increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of steroid hormones,compounds with a 1, 2, cyclopentanoperhydrophenanthrene nucleus that act as hormones. |
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| prostate gland epithelium morphogenesis | The process in which the anatomical structures of epithelia of the prostate gland are generated and organized. An epithelium consists of closely packed cells arranged in one or more layers, that covers the outer surfaces of the body or lines any internal cavity or tube. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of JNK cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the JNK cascade. |
| response to ethanol | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ethanol stimulus. |
| response to L-glutamate | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an L-glutamate stimulus. |
| transcytosis | The directed movement of endocytosed material through the cell and its exocytosis from the plasma membrane at the opposite side. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
70 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q05688 | IGF1R | Insulin-like growth factor 1 receptor | Bos taurus (Bovine) | SS |
| P09208 | InR | Insulin-like receptor | Drosophila melanogaster (Fruit fly) | SS |
| P06213 | INSR | Insulin receptor | Homo sapiens (Human) | EV |
| P14616 | INSRR | Insulin receptor-related protein | Homo sapiens (Human) | SS |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | Homo sapiens (Human) | EV |
| P35546 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Mus musculus (Mouse) | SS |
| Q61006 | Musk | Muscle, skeletal receptor tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q9Z138 | Ror2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Mus musculus (Mouse) | SS |
| Q9Z139 | Ror1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Mus musculus (Mouse) | PR |
| Q62371 | Ddr2 | Discoidin domain-containing receptor 2 | Mus musculus (Mouse) | SS |
| Q03146 | Ddr1 | Epithelial discoidin domain-containing receptor 1 | Mus musculus (Mouse) | SS |
| P15209 | Ntrk2 | BDNF/NT-3 growth factors receptor | Mus musculus (Mouse) | SS |
| Q3UFB7 | Ntrk1 | High affinity nerve growth factor receptor | Mus musculus (Mouse) | SS |
| Q6VNS1 | Ntrk3 | NT-3 growth factor receptor | Mus musculus (Mouse) | SS |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P35969 | Flt1 | Vascular endothelial growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P97793 | Alk | ALK tyrosine kinase receptor | Mus musculus (Mouse) | SS |
| Q9WTL4 | Insrr | Insulin receptor-related protein | Mus musculus (Mouse) | SS |
| P15208 | Insr | Insulin receptor | Mus musculus (Mouse) | SS |
| P16092 | Fgfr1 | Fibroblast growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P21803 | Fgfr2 | Fibroblast growth factor receptor 2 | Mus musculus (Mouse) | SS |
| Q03142 | Fgfr4 | Fibroblast growth factor receptor 4 | Mus musculus (Mouse) | PR |
| Q60805 | Mertk | Tyrosine-protein kinase Mer | Mus musculus (Mouse) | SS |
| Q62190 | Mst1r | Macrophage-stimulating protein receptor | Mus musculus (Mouse) | SS |
| Q00993 | Axl | Tyrosine-protein kinase receptor UFO | Mus musculus (Mouse) | PR |
| P55144 | Tyro3 | Tyrosine-protein kinase receptor TYRO3 | Mus musculus (Mouse) | SS |
| P70424 | Erbb2 | Receptor tyrosine-protein kinase erbB-2 | Mus musculus (Mouse) | SS |
| Q61527 | Erbb4 | Receptor tyrosine-protein kinase erbB-4 | Mus musculus (Mouse) | SS |
| Q61526 | Erbb3 | Receptor tyrosine-protein kinase erbB-3 | Mus musculus (Mouse) | SS |
| Q01279 | Egfr | Epidermal growth factor receptor | Mus musculus (Mouse) | SS |
| Q01887 | Ryk | Tyrosine-protein kinase RYK | Mus musculus (Mouse) | PR |
| Q02858 | Tek | Angiopoietin-1 receptor | Mus musculus (Mouse) | SS |
| Q06806 | Tie1 | Tyrosine-protein kinase receptor Tie-1 | Mus musculus (Mouse) | SS |
| P05532 | Kit | Mast/stem cell growth factor receptor Kit | Mus musculus (Mouse) | PR |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| P09581 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Mus musculus (Mouse) | SS |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| Q00342 | Flt3 | Receptor-type tyrosine-protein kinase FLT3 | Mus musculus (Mouse) | SS |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| Q61851 | Fgfr3 | Fibroblast growth factor receptor 3 | Mus musculus (Mouse) | PR |
| Q64716 | Insrr | Insulin receptor-related protein | Rattus norvegicus (Rat) | SS |
| P15127 | Insr | Insulin receptor | Rattus norvegicus (Rat) | SS |
| P24062 | Igf1r | Insulin-like growth factor 1 receptor | Rattus norvegicus (Rat) | SS |
| O64770 | At1g61490 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61490 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64783 | At1g61370 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81833 | SD11 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LPZ9 | SD113 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LW83 | CES101 | G-type lectin S-receptor-like serine/threonine-protein kinase CES101 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SXB5 | At1g11303 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11303 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SYA0 | At1g61500 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61500 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64477 | At2g19130 | G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SXB4 | At1g11300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64778 | At1g61420 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61420 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64782 | SD129 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-29 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64774 | At1g61460 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61460 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SY95 | At1g61550 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61550 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64780 | At1g61400 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61400 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64776 | At1g61440 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61440 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64793 | At1g67520 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39203 | SD22 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64784 | At1g61360 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61360 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKSGSGGGSP | TSLWGLVFLS | AALSLWPTSG | EICGPGIDIR | NDYQQLKRLE | NCTVIEGFLH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ILLISKAEDY | RSYRFPKLTV | ITEYLLLFRV | AGLESLGDLF | PNLTVIRGWK | LFYNYALVIF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EMTNLKDIGL | YNLRNITRGA | IRIEKNADLC | YLSTIDWSLI | LDAVSNNYIV | GNKPPKECGD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LCPGTLEEKP | MCEKTTINNE | YNYRCWTTNR | CQKMCPSVCG | KRACTENNEC | CHPECLGSCH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TPDDNTTCVA | CRHYYYKGVC | VPACPPGTYR | FEGWRCVDRD | FCANIPNAES | SDSDGFVIHD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DECMQECPSG | FIRNSTQSMY | CIPCEGPCPK | VCGDEEKKTK | TIDSVTSAQM | LQGCTILKGN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LLINIRRGNN | IASELENFMG | LIEVVTGYVK | IRHSHALVSL | SFLKNLRLIL | GEEQLEGNYS |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FYVLDNQNLQ | QLWDWNHRNL | TVRSGKMYFA | FNPKLCVSEI | YRMEEVTGTK | GRQSKGDINT |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RNNGERASCE | SDVLRFTSTT | TWKNRIIITW | HRYRPPDYRD | LISFTVYYKE | APFKNVTEYD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GQDACGSNSW | NMVDVDLPPN | KEGEPGILLH | GLKPWTQYAV | YVKAVTLTMV | ENDHIRGAKS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EILYIRTNAS | VPSIPLDVLS | ASNSSSQLIV | KWNPPTLPNG | NLSYYIVRWQ | RQPQDGYLYR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HNYCSKDKIP | IRKYADGTID | VEEVTENPKT | EVCGGDKGPC | CACPKTEAEK | QAEKEEAEYR |
| 730 | 740 | 750 | 760 | 770 | 780 |
| KVFENFLHNS | IFVPRPERRR | RDVMQVANTT | MSSRSRNTTV | ADTYNITDPE | EFETEYPFFE |
| 790 | 800 | 810 | 820 | 830 | 840 |
| SRVDNKERTV | ISNLRPFTLY | RIDIHSCNHE | AEKLGCSASN | FVFARTMPAE | GADDIPGPVT |
| 850 | 860 | 870 | 880 | 890 | 900 |
| WEPRPENSIF | LKWPEPENPN | GLILMYEIKY | GSQVEDQREC | VSRQEYRKYG | GAKLNRLNPG |
| 910 | 920 | 930 | 940 | 950 | 960 |
| NYTARIQATS | LSGNGSWTDP | VFFYVPAKTT | YENFMHLIIA | LPVAILLIVG | GLVIMLYVFH |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| RKRNNSRLGN | GVLYASVNPE | YFSAADVYVP | DEWEVAREKI | TMNRELGQGS | FGMVYEGVAK |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| GVVKDEPETR | VAIKTVNEAA | SMRERIEFLN | EASVMKEFNC | HHVVRLLGVV | SQGQPTLVIM |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| ELMTRGDLKS | YLRSLRPEVE | QNNLVLIPPS | LSKMIQMAGE | IADGMAYLNA | NKFVHRDLAA |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| RNCMVAEDFT | VKIGDFGMTR | DIYETDYYRK | GGKGLLPVRW | MSPESLKDGV | FTTHSDVWSF |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| GVVLWEIATL | AEQPYQGLSN | EQVLRFVMEG | GLLDKPDNCP | DMLFELMRMC | WQYNPKMRPS |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| FLEIIGSIKD | EMEPSFQEVS | FYYSEENKPP | EPEELEMELE | MEPENMESVP | LDPSASSASL |
| 1330 | 1340 | 1350 | 1360 | 1370 | |
| PLPERHSGHK | AENGPGPGVL | VLRASFDERQ | PYAHMNGGRA | NERALPLPQS | STC |