Q5RJQ4
Gene name |
Sirt2 (Sir2l2) |
Protein name |
NAD-dependent protein deacetylase sirtuin-2 |
Names |
NAD-dependent protein defatty-acylase sirtuin-2, Regulatory protein SIR2 homolog 2, SIR2-like protein 2 |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:361532 |
EC number |
2.3.1.286: Transferring groups other than amino-acyl groups |
Protein Class |
|
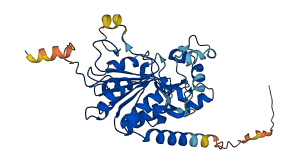
Descriptions
(Annotation based on sequence homology with Q8IXJ6)
Sirtuin 2 (SIRT2) is a NAD+ dependent deacetylase that has been associated with neurodegeneration and cancer. The C-terminal region (CT) of SIRT2 functions as an autoinhibitory region that regulates the deacetylation activity of SIRT2. Phosphorylation at S331 of SIRT2 isoform 2 causes large conformational changes in the CT that enhance the autoinhibitory activity of the CT region. This serine residue is located within the naturally disordered C-terminal region (CT, residues 320-352 in isoform 2).
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q5RJQ4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5RJQ4-F1 | Predicted | AlphaFoldDB |
No variants for Q5RJQ4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q5RJQ4 | |||||
No associated diseases with Q5RJQ4
1 regional properties for Q5RJQ4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Sirtuin family, catalytic core domain | 20 - 301 | IPR026590 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.1.286 | Transferring groups other than amino-acyl groups |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
27 GO annotations of cellular component
| Name | Definition |
|---|---|
| centriole | A cellular organelle, found close to the nucleus in many eukaryotic cells, consisting of a small cylinder with microtubular walls, 300-500 nm long and 150-250 nm in diameter. It contains nine short, parallel, peripheral microtubular fibrils, each fibril consisting of one complete microtubule fused to two incomplete microtubules. Cells usually have two centrioles, lying at right angles to each other. At division, each pair of centrioles generates another pair and the twin pairs form the pole of the mitotic spindle. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| chromosome | A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| glial cell projection | A prolongation or process extending from a glial cell. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| heterochromatin | A compact and highly condensed form of chromatin that is refractory to transcription. |
| juxtaparanode region of axon | A region of an axon near a node of Ranvier that is between the paranode and internode regions. |
| lateral loop | Non-compact myelin located adjacent to the nodes of Ranvier in a myelin segment. These non-compact regions include cytoplasm from the cell responsible for synthesizing the myelin. Lateral loops are found in the paranodal region adjacent to the nodes of Ranvier, while Schmidt-Lantermann clefts are analogous structures found within the compact myelin internode. |
| meiotic spindle | A spindle that forms as part of meiosis. Several proteins, such as budding yeast Spo21p, fission yeast Spo2 and Spo13, and C. elegans mei-1, localize specifically to the meiotic spindle and are absent from the mitotic spindle. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| mitotic spindle | A spindle that forms as part of mitosis. Mitotic and meiotic spindles contain distinctive complements of proteins associated with microtubules. |
| myelin sheath | An electrically insulating fatty layer that surrounds the axons of many neurons. It is an outgrowth of glial cells: Schwann cells supply the myelin for peripheral neurons while oligodendrocytes supply it to those of the central nervous system. |
| myelin sheath abaxonal region | The region of the myelin sheath furthest from the axon. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| paranodal junction | A highly specialized cell-cell junction found in vertebrates, which forms between a neuron and a glial cell, and has structural similarity to Drosophila septate junctions. It flanks the node of Ranvier in myelinated nerve and electrically isolates the myelinated from unmyelinated nerve segments and physically separates the voltage-gated sodium channels at the node from the cluster of potassium channels underneath the myelin sheath. |
| paranode region of axon | An axon part that is located adjacent to the nodes of Ranvier and surrounded by lateral loop portions of myelin sheath. |
| perikaryon | The portion of the cell soma (neuronal cell body) that excludes the nucleus. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| Schmidt-Lanterman incisure | Regions within compact myelin in which the cytoplasmic faces of the enveloping myelin sheath are not tightly juxtaposed, and include cytoplasm from the cell responsible for making the myelin. Schmidt-Lanterman incisures occur in the compact myelin internode, while lateral loops are analogous structures found in the paranodal region adjacent to the nodes of Ranvier. |
| spindle | The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart. |
| terminal loop | Portion of myelin-forming Schwann cell consisting of terminal cytoplasmic extensions adhered to the axon at the beginning and end of the myelin sheath. |
17 GO annotations of molecular function
| Name | Definition |
|---|---|
| beta-tubulin binding | Binding to the microtubule constituent protein beta-tubulin. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| histone acetyltransferase binding | Binding to an histone acetyltransferase. |
| histone deacetylase activity | Catalysis of the reaction: histone N6-acetyl-L-lysine + H2O = histone L-lysine + acetate. This reaction represents the removal of an acetyl group from a histone, a class of proteins complexed to DNA in chromatin and chromosomes. |
| histone deacetylase binding | Binding to histone deacetylase. |
| NAD+ ADP-ribosyltransferase activity | Catalysis of the reaction: NAD+ + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor. |
| NAD+ binding | Binding to the oxidized form, NAD, of nicotinamide adenine dinucleotide, a coenzyme involved in many redox and biosynthetic reactions. |
| NAD-dependent histone deacetylase activity | Catalysis of the reaction: histone N6-acetyl-L-lysine + H2O = histone L-lysine + acetate. This reaction requires the presence of NAD, and represents the removal of an acetyl group from a histone. |
| NAD-dependent histone deacetylase activity (H4-K16 specific) | Catalysis of the reaction: histone H4 N6-acetyl-L-lysine (position 16) + H2O = histone H4 L-lysine (position 16) + acetate. This reaction requires the presence of NAD, and represents the removal of an acetyl group from lysine at position 16 of the histone H4 protein. |
| NAD-dependent protein deacetylase activity | Catalysis of the removal of one or more acetyl groups from a protein, requiring NAD. |
| NAD-dependent protein demyristoylase activity | Catalysis of the reaction: N6-tetradecanoyl-L-lysyl- + NAD(+) + H2O = tetradecanoyl-ADP-D-ribose + L-lysyl- |
| NAD-dependent protein depalmitoylase activity | Catalysis of the reaction: N6-octadecanoyl-L-lysyl- + NAD(+) + H2O = octadecanoyl-ADP-D-ribose + L-lysyl- |
| protein lysine deacetylase activity | Catalysis of the reaction: H2O + N6-acetyl-L-lysyl- |
| tubulin deacetylase activity | Catalysis of the reaction: N-acetyl(alpha-tubulin) + H2O = alpha-tubulin + acetate. |
| ubiquitin binding | Binding to ubiquitin, a protein that when covalently bound to other cellular proteins marks them for proteolytic degradation. |
| zinc ion binding | Binding to a zinc ion (Zn). |
52 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| cellular lipid catabolic process | The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells. |
| cellular response to caloric restriction | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a of caloric restriction, insufficient food energy intake. |
| cellular response to epinephrine stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an epinephrine stimulus. Epinephrine is a catecholamine that has the formula C9H13NO3; it is secreted by the adrenal medulla to act as a hormone, and released by certain neurons to act as a neurotransmitter active in the central nervous system. |
| cellular response to hepatocyte growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hepatocyte growth factor stimulus. |
| cellular response to hypoxia | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| cellular response to molecule of bacterial origin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin. |
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| central nervous system development | The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain and spinal cord. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| hepatocyte growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a hepatocyte growth factor receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| histone deacetylation | The modification of histones by removal of acetyl groups. |
| histone H3 deacetylation | The modification of histone H3 by the removal of one or more acetyl groups. |
| histone H4 deacetylation | The modification of histone H4 by the removal of one or more acetyl groups. |
| meiotic cell cycle | Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell via two nuclear divisions. |
| myelination in peripheral nervous system | The process in which neuronal axons and dendrites become coated with a segmented lipid-rich sheath (myelin) to enable faster and more energetically efficient conduction of electrical impulses. The sheath is formed by the cell membranes of Schwann cells in the peripheral nervous system. Adjacent myelin segments are separated by a non-myelinated stretch of axon called a node of Ranvier. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of autophagy | Any process that stops, prevents, or reduces the frequency, rate or extent of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of defense response to bacterium | Any process that stops, prevents or reduces the frequency, rate or extent of defense response to bacterium. |
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| negative regulation of fat cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of adipocyte differentiation. |
| negative regulation of NLRP3 inflammasome complex assembly | Any process that stops, prevents or reduces the frequency, rate or extent of NLRP3 inflammasome complex assembly. |
| negative regulation of oligodendrocyte differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of oligodendrocyte differentiation. |
| negative regulation of oligodendrocyte progenitor proliferation | Any process that stops or decreases the rate or extent of oligodendrocyte progenitor proliferation. |
| negative regulation of peptidyl-threonine phosphorylation | Any process that decreases the frequency, rate or extent of peptidyl-threonine phosphorylation. Peptidyl-threonine phosphorylation is the phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
| negative regulation of protein catabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of protein catabolic process. |
| negative regulation of reactive oxygen species metabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of reactive oxygen species metabolic process. |
| negative regulation of striated muscle tissue development | Any process that stops, prevents, or reduces the frequency, rate or extent of striated muscle development. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | Any process that decreases the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of a hypoxia stimulus. |
| NLRP3 inflammasome complex assembly | The aggregation, arrangement and bonding together of a set of components to form the NLRP3 inflammasome complex, occurring at the level of an individual cell. |
| peptidyl-lysine deacetylation | The removal of an acetyl group from an acetylated lysine residue in a peptide or protein. |
| phosphatidylinositol 3-kinase signaling | A series of reactions within the signal-receiving cell, mediated by the intracellular phosphatidylinositol 3-kinase (PI3K). Many cell surface receptor linked signaling pathways signal through PI3K to regulate numerous cellular functions. |
| positive regulation of attachment of spindle microtubules to kinetochore | Any process that activates or increases the frequency, rate or extent of the attachment of spindle microtubules to the kinetochore. |
| positive regulation of cell division | Any process that activates or increases the frequency, rate or extent of cell division. |
| positive regulation of DNA binding | Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of execution phase of apoptosis | Any process that activates or increases the frequency, rate or extent of execution phase of apoptosis. |
| positive regulation of meiotic nuclear division | Any process that activates or increases the frequency, rate or extent of meiosis. |
| positive regulation of oocyte maturation | Any process that activates or increases the frequency, rate or extent of oocyte maturation. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | Any positive regulation of proteasomal ubiquitin-dependent protein catabolic process that is involved in a cellular response to hypoxia. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| proteasome-mediated ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein ADP-ribosylation | The transfer, from NAD, of ADP-ribose to protein amino acids. |
| protein deacetylation | The removal of an acetyl group from a protein amino acid. An acetyl group is CH3CO-, derived from acetic |
| protein kinase B signaling | A series of reactions, mediated by the intracellular serine/threonine kinase protein kinase B (also called AKT), which occurs as a result of a single trigger reaction or compound. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of fat cell differentiation | Any process that modulates the frequency, rate or extent of adipocyte differentiation. |
| regulation of myelination | Any process that modulates the frequency, rate or extent of the formation of a myelin sheath around nerve axons. |
| regulation of postsynaptic neurotransmitter receptor internalization | Any process that modulates the frequency, rate or extent of endocytosis of neurotransmitter receptor at the postsynapse. |
| tubulin deacetylation | The removal of an acetyl group from tubulin. An acetyl group is CH3CO-, derived from acetic |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P53686 | HST2 | NAD-dependent protein deacetylase HST2 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| Q9I7I7 | Sirt2 | NAD-dependent protein deacetylase Sirt2 | Drosophila melanogaster (Fruit fly) | SS |
| Q9NTG7 | SIRT3 | NAD-dependent protein deacetylase sirtuin-3, mitochondrial | Homo sapiens (Human) | SS |
| Q8IXJ6 | SIRT2 | NAD-dependent protein deacetylase sirtuin-2 | Homo sapiens (Human) | EV |
| Q8R104 | Sirt3 | NAD-dependent protein deacetylase sirtuin-3 | Mus musculus (Mouse) | SS |
| Q8VDQ8 | Sirt2 | NAD-dependent protein deacetylase sirtuin-2 | Mus musculus (Mouse) | SS |
| Q7ZVK3 | sirt2 | NAD-dependent protein deacetylase sirtuin-2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDFLRNLFTQ | TLGLGSQKER | LLDELTLEGV | TRYMQSERCR | RVICLVGAGI | STSAGIPDFR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SPSTGLYANL | EKYHLPYPEA | IFEISYFKKH | PEPFFALAKE | LYPGQFKPTI | CHYFIRLLKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KGLLLRCYTQ | NIDTLERVAG | LEPQDLVEAH | GTFYTSHCVN | TSCGKEYTMS | WMKEKIFSEA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TPKCEKCQNV | VKPDIVFFGE | NLPPRFFSCM | QSDFSKVDLL | IIMGTSLQVQ | PFASLISKAP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LATPRLLINK | EKTGQTDPFL | GMMMGLGGGM | DFDSKKAYRD | VAWLGDCDQG | CLALADLLGW |
| 310 | 320 | 330 | 340 | ||
| KELEDLVRRE | HANIDAQSGS | QASNPSATVS | PRKSPPPAKE | AARTKEKEEH |