Q5K5B6
Gene name |
Os07g0647900 (LOC_Os07g45360, OJ1316_A04.119, OsJ_25369, P0427D10.102) |
Protein name |
DEAD-box ATP-dependent RNA helicase 57 |
Names |
|
Species |
Oryza sativa subsp japonica (Rice) |
KEGG Pathway |
osa:4344113 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
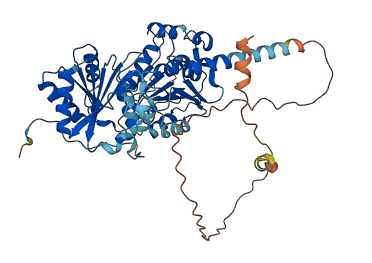
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q5K5B6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5K5B6-F1 | Predicted | AlphaFoldDB |
No variants for Q5K5B6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q5K5B6 | |||||
No associated diseases with Q5K5B6
5 regional properties for Q5K5B6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Helicase, C-terminal | 375 - 519 | IPR001650 |
| domain | DEAD/DEAH box helicase domain | 170 - 335 | IPR011545 |
| domain | Helicase superfamily 1/2, ATP-binding domain | 165 - 362 | IPR014001 |
| domain | RNA helicase, DEAD-box type, Q motif | 146 - 174 | IPR014014 |
| domain | DDX52/Rok1, DEAD-box helicase domain | 157 - 350 | IPR044764 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| maturation of SSU-rRNA | Any process involved in the maturation of a precursor Small SubUnit (SSU) ribosomal RNA (rRNA) molecule into a mature SSU-rRNA molecule. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P45818 | ROK1 | ATP-dependent RNA helicase ROK1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q6Z2Z4 | Os02g0146600 | Eukaryotic initiation factor 4A-3 | Oryza sativa subsp japonica (Rice) | PR |
| P35683 | Os06g0701100 | Eukaryotic initiation factor 4A-1 | Oryza sativa subsp japonica (Rice) | PR |
| Q7Y183 | RH10 | DEAD-box ATP-dependent RNA helicase 10 | Oryza sativa subsp japonica (Rice) | PR |
| Q8L4E9 | Os07g0633500 | DEAD-box ATP-dependent RNA helicase 36 | Oryza sativa subsp japonica (Rice) | PR |
| Q0DBS1 | Os06g0535100 | Putative DEAD-box ATP-dependent RNA helicase 51 | Oryza sativa subsp japonica (Rice) | PR |
| Q0D622 | Os07g0517000 | DEAD-box ATP-dependent RNA helicase 32 | Oryza sativa subsp japonica (Rice) | PR |
| Q84TG1 | RH57 | DEAD-box ATP-dependent RNA helicase 57 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEKAKLSSAL | FAGTHFDRKR | FAGDFARFRQ | GPPAPDVASA | AAPSPEKKRK | RQSKAKAKKS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KKRRAEGADS | ASDAVEGFSV | FKGLAAKKDE | DDSEKKVETG | KSEDSEVVRR | RKEVEREIER |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AAILRKKFDI | HISGQNVPAP | LENFEELVSR | YGCDSYLVGN | LSKLGFQEPT | PIQRQAIPIL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSGRECFACA | PTGSGKTLAF | LFPILMKIKP | GSKEGVKAVI | LCPTRELAAQ | TTRECKKLAK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GRKFYIKLMT | KDLSKSGNFK | DMHCDILIST | PLRLDHAVQK | RDLDLSRVEY | LVLDESDKLF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ELGFVEVIDS | VVKACSNPSI | IRSLFSATLP | DSIETLARTI | MHDAVRVIVG | RKNSASSLIK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QKLIFAGTEK | GKLLALRQSF | AESLNPPVLI | FVQSKERAKE | LYKELAFDDV | RADVIHADLD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| EEQRQDAVDN | LRAGKTWVLI | ATEVIARGMD | FKGVNCVINY | DFPESASAYI | HRIGRSGRAG |
| 490 | 500 | 510 | 520 | 530 | |
| RSGEAITFFT | EEDKPFLRNI | ANVLISSGCE | VPSWIKALPK | LKRKKHRVNR | DPISTLPDED |