Q5G5E0
Gene name |
PIRL5 (At2g17440, F5J6.20) |
Protein name |
Plant intracellular Ras-group-related LRR protein 5 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT2G17440 |
EC number |
|
Protein Class |
|
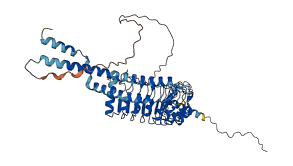
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q5G5E0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5G5E0-F1 | Predicted | AlphaFoldDB |
53 variants for Q5G5E0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH05513949 | 7 | V>I | No | 1000Genomes | |
| ENSVATH01827357 | 14 | I>M | No | 1000Genomes | |
| ENSVATH00234653 | 20 | L>S | No | 1000Genomes | |
| ENSVATH01827359 | 23 | E>D | No | 1000Genomes | |
| ENSVATH01827360 | 25 | D>E | No | 1000Genomes | |
| tmp_2_7571422_C_A | 31 | T>N | No | 1000Genomes | |
| tmp_2_7571443_A_G | 38 | E>G | No | 1000Genomes | |
| ENSVATH01827361 | 38 | E>Q | No | 1000Genomes | |
| ENSVATH00234657 | 45 | L>F | No | 1000Genomes | |
| ENSVATH00234658 | 48 | I>N | No | 1000Genomes | |
| ENSVATH05513951 | 53 | K>I | No | 1000Genomes | |
| ENSVATH14509588 | 54 | T>I | No | 1000Genomes | |
| ENSVATH00234661 | 67 | F>L | No | 1000Genomes | |
| tmp_2_7571548_G_A | 73 | S>N | No | 1000Genomes | |
| ENSVATH05513952 | 77 | F>Y | No | 1000Genomes | |
| ENSVATH05513953 | 83 | T>R | No | 1000Genomes | |
| tmp_2_7571631_G_A | 101 | E>K | No | 1000Genomes | |
| ENSVATH00234662 | 111 | A>V | No | 1000Genomes | |
| ENSVATH13236586 | 113 | P>H | No | 1000Genomes | |
| ENSVATH01827362 | 115 | S>G | No | 1000Genomes | |
| ENSVATH01827363 | 115 | S>T | No | 1000Genomes | |
| ENSVATH05513954 | 118 | A>D | No | 1000Genomes | |
| ENSVATH01827364 | 145 | R>H | No | 1000Genomes | |
| ENSVATH13236587 | 169 | Q>H | No | 1000Genomes | |
| tmp_2_7572259_A_T | 184 | N>Y | No | 1000Genomes | |
| tmp_2_7572298_T_G | 197 | L>V | No | 1000Genomes | |
| ENSVATH14509594 | 218 | Q>P | No | 1000Genomes | |
| ENSVATH13236593 | 221 | W>* | No | 1000Genomes | |
| ENSVATH13236594 | 226 | L>I | No | 1000Genomes | |
| ENSVATH13236605 | 229 | L>I | No | 1000Genomes | |
| tmp_2_7572415_G_C | 236 | D>H | No | 1000Genomes | |
| ENSVATH13236609 | 249 | I>V | No | 1000Genomes | |
| ENSVATH13236611 | 253 | I>F | No | 1000Genomes | |
| ENSVATH13236612 | 293 | S>A | No | 1000Genomes | |
| ENSVATH00234666 | 295 | F>L | No | 1000Genomes | |
| tmp_2_7572640_C_T | 311 | L>F | No | 1000Genomes | |
| ENSVATH13236614 | 312 | S>T | No | 1000Genomes | |
| ENSVATH05513970 | 325 | K>R | No | 1000Genomes | |
| tmp_2_7572851_G_A | 381 | R>Q | No | 1000Genomes | |
| tmp_2_7572883_A_C | 392 | N>H | No | 1000Genomes | |
| ENSVATH05513971 | 393 | L>I | No | 1000Genomes | |
| tmp_2_7572932_A_G | 408 | E>G | No | 1000Genomes | |
| ENSVATH13236627 | 413 | A>V | No | 1000Genomes | |
| tmp_2_7573011_A_C | 434 | L>F | No | 1000Genomes | |
| tmp_2_7573064_G_A | 452 | R>H | No | 1000Genomes | |
| ENSVATH01827368 | 457 | S>T | No | 1000Genomes | |
| ENSVATH14509616 | 458 | F>V | No | 1000Genomes | |
| tmp_2_7573144_A_G | 479 | R>G | No | 1000Genomes | |
| ENSVATH05513976 | 497 | V>M | No | 1000Genomes | |
| ENSVATH14509617 | 503 | K>R | No | 1000Genomes | |
| tmp_2_7573378_G_A | 518 | C>Y | No | 1000Genomes | |
| ENSVATH05513977 | 522 | K>E | No | 1000Genomes | |
| ENSVATH13236628 | 523 | S>P | No | 1000Genomes |
No associated diseases with Q5G5E0
13 regional properties for Q5G5E0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 223 - 275 | IPR001611-1 |
| repeat | Leucine-rich repeat | 277 - 334 | IPR001611-2 |
| repeat | Leucine-rich repeat | 347 - 413 | IPR001611-3 |
| repeat | Leucine-rich repeat | 440 - 461 | IPR001611-4 |
| repeat | Leucine-rich repeat, typical subtype | 229 - 251 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 252 - 274 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 275 - 297 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 298 - 320 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 321 - 344 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 367 - 389 | IPR003591-6 |
| repeat | Leucine-rich repeat, typical subtype | 390 - 413 | IPR003591-7 |
| repeat | Leucine-rich repeat, typical subtype | 438 - 460 | IPR003591-8 |
| repeat | Leucine-rich repeat, typical subtype | 461 - 484 | IPR003591-9 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
40 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3MHH9 | ECM2 | Extracellular matrix protein 2 | Bos taurus (Bovine) | PR |
| P58874 | OPTC | Opticin | Bos taurus (Bovine) | PR |
| Q24K06 | LRRC10 | Leucine-rich repeat-containing protein 10 | Bos taurus (Bovine) | PR |
| Q9V780 | Lap1 | Protein lap1 | Drosophila melanogaster (Fruit fly) | PR |
| Q96NW7 | LRRC7 | Leucine-rich repeat-containing protein 7 | Homo sapiens (Human) | PR |
| Q8IWT6 | LRRC8A | Volume-regulated anion channel subunit LRRC8A | Homo sapiens (Human) | PR |
| Q9HCJ2 | LRRC4C | Leucine-rich repeat-containing protein 4C | Homo sapiens (Human) | PR |
| Q9UFC0 | LRWD1 | Leucine-rich repeat and WD repeat-containing protein 1 | Homo sapiens (Human) | PR |
| Q86UN2 | RTN4RL1 | Reticulon-4 receptor-like 1 | Homo sapiens (Human) | PR |
| Q8IWK6 | ADGRA3 | Adhesion G protein-coupled receptor A3 | Homo sapiens (Human) | PR |
| Q7L1W4 | LRRC8D | Volume-regulated anion channel subunit LRRC8D | Homo sapiens (Human) | PR |
| Q96FE5 | LINGO1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Homo sapiens (Human) | PR |
| Q8TDW0 | LRRC8C | Volume-regulated anion channel subunit LRRC8C | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| Q96L50 | LRR1 | Leucine-rich repeat protein 1 | Homo sapiens (Human) | PR |
| A6H694 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Mus musculus (Mouse) | PR |
| Q7TT36 | Adgra3 | Adhesion G protein-coupled receptor A3 | Mus musculus (Mouse) | PR |
| P59383 | Lrrn4 | Leucine-rich repeat neuronal protein 4 | Mus musculus (Mouse) | PR |
| Q9D1T0 | Lingo1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Mus musculus (Mouse) | PR |
| Q8R502 | Lrrc8c | Volume-regulated anion channel subunit LRRC8C | Mus musculus (Mouse) | PR |
| Q80WG5 | Lrrc8a | Volume-regulated anion channel subunit LRRC8A | Mus musculus (Mouse) | PR |
| Q8K0S5 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Mus musculus (Mouse) | PR |
| Q80TE7 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Mus musculus (Mouse) | PR |
| Q5DU41 | Lrrc8b | Volume-regulated anion channel subunit LRRC8B | Mus musculus (Mouse) | PR |
| Q5RKR3 | Islr2 | Immunoglobulin superfamily containing leucine-rich repeat protein 2 | Mus musculus (Mouse) | PR |
| Q8C031 | Lrrc4c | Leucine-rich repeat-containing protein 4C | Mus musculus (Mouse) | PR |
| Q8BGI7 | Lrrc39 | Leucine-rich repeat-containing protein 39 | Mus musculus (Mouse) | PR |
| Q9D9Q0 | Lrrc69 | Leucine-rich repeat-containing protein 69 | Mus musculus (Mouse) | PR |
| P70587 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Rattus norvegicus (Rat) | PR |
| Q4V8G0 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Rattus norvegicus (Rat) | PR |
| Q80WD0 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Rattus norvegicus (Rat) | PR |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9SHI4 | RLP3 | Receptor-like protein 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22727 | At1g61190 | Probable disease resistance protein At1g61190 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P60838 | SUMM2 | Disease resistance protein SUMM2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64790 | At1g61300 | Probable disease resistance protein At1g61300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64973 | RPS5 | Disease resistance protein RPS5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| B0JZ65 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q68F79 | lrrc8e | Volume-regulated anion channel subunit LRRC8E | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| B0R160 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDSKEMVVEE | IMRIHRSLPL | RPEIDDVETA | TSLIQNVEKE | DRNRLEAIDK | LVKTSSSEVP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LELFNVFKEM | KKSLVRFQST | EQTREATKIL | DLESVHVVFD | ELIQRASFCI | ASPNSTTALP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RSVPVPAPVV | SSDEIPFKSK | EIISRDDTFV | KKAKSSFYSD | GLLAPSKPQV | LDSTLHQAKN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VAGNDGEKLS | LIKLASLIEV | SAKKATQELN | LQHRLMDQLE | WLPDSLGKLS | SLVRLDLSEN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| CIMVLPATIG | GLISLTRLDL | HSNRIGQLPE | SIGDLLNLVN | LNLSGNQLSS | LPSSFNRLIH |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LEELDLSSNS | LSILPESIGS | LVSLKKLDVE | TNNIEEIPHS | ISGCSSMEEL | RADYNRLKAL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PEAVGKLSTL | EILTVRYNNI | RQLPTTMSSM | ANLKELDVSF | NELESVPESL | CYAKTLVKLN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| IGNNFANLRS | LPGLIGNLEK | LEELDMSNNQ | IRFLPYSFKT | LSNLRVLQTE | QNPLEELPRD |
| 490 | 500 | 510 | 520 | ||
| ITEKGAQAVV | QYMNDLVEAR | NTKSQRTKPK | KSWVNSICFF | CKSSTN |