Q5F470
Gene name |
RAB8A (RCJMB04_2k8) |
Protein name |
Ras-related protein Rab-8A |
Names |
|
Species |
Gallus gallus (Chicken) |
KEGG Pathway |
gga:428352 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
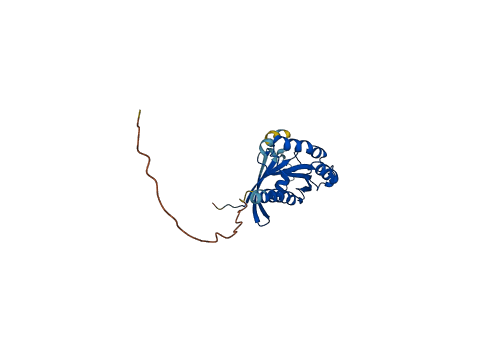
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q5F470
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5F470-F1 | Predicted | AlphaFoldDB |
No variants for Q5F470
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q5F470 | |||||
No associated diseases with Q5F470
1 regional properties for Q5F470
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 7 - 162 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
| trans-Golgi network transport vesicle | A vesicle that mediates transport between the trans-Golgi network and other parts of the cell. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| small GTPase binding | Binding to a small monomeric GTPase. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| axonogenesis | De novo generation of a long process of a neuron, including the terminal branched region. Refers to the morphogenesis or creation of shape or form of the developing axon, which carries efferent (outgoing) action potentials from the cell body towards target cells. |
| cilium assembly | The assembly of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole. |
| endocytic recycling | The directed movement of membrane-bounded vesicles from endosomes back to the plasma membrane, a trafficking pathway that promotes the recycling of internalized transmembrane proteins. |
| Golgi organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the Golgi apparatus. |
| Golgi vesicle fusion to target membrane | The joining of the lipid bilayer membrane around a Golgi transport vesicle to the target lipid bilayer membrane. |
| neurotransmitter receptor transport to postsynaptic membrane | The directed movement of neurotransmitter receptor to the postsynaptic membrane in transport vesicles. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein secretion | The controlled release of proteins from a cell. |
| regulation of exocytosis | Any process that modulates the frequency, rate or extent of exocytosis. |
| vesicle docking involved in exocytosis | The initial attachment of a vesicle membrane to a target membrane, mediated by proteins protruding from the membrane of the vesicle and the target membrane, that contributes to exocytosis. |
28 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P07560 | SEC4 | Ras-related protein SEC4 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q2HJI8 | RAB8B | Ras-related protein Rab-8B | Bos taurus (Bovine) | PR |
| Q1RMR4 | RAB15 | Ras-related protein Rab-15 | Bos taurus (Bovine) | PR |
| A4FV54 | RAB8A | Ras-related protein Rab-8A | Bos taurus (Bovine) | PR |
| P61007 | RAB8A | Ras-related protein Rab-8A | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P0C0E4 | RAB40AL | Ras-related protein Rab-40A-like | Homo sapiens (Human) | PR |
| Q92930 | RAB8B | Ras-related protein Rab-8B | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q8WXH6 | RAB40A | Ras-related protein Rab-40A | Homo sapiens (Human) | PR |
| Q96S21 | RAB40C | Ras-related protein Rab-40C | Homo sapiens (Human) | PR |
| Q12829 | RAB40B | Ras-related protein Rab-40B | Homo sapiens (Human) | PR |
| P61006 | RAB8A | Ras-related protein Rab-8A | Homo sapiens (Human) | PR |
| Q8K386 | Rab15 | Ras-related protein Rab-15 | Mus musculus (Mouse) | PR |
| P61028 | Rab8b | Ras-related protein Rab-8B | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q9DD03 | Rab13 | Ras-related protein Rab-13 | Mus musculus (Mouse) | PR |
| Q8CB87 | Rab44 | Ras-related protein Rab-44 | Mus musculus (Mouse) | PR |
| Q8VHP8 | Rab40b | Ras-related protein Rab-40B | Mus musculus (Mouse) | PR |
| Q8VHQ4 | Rab40c | Ras-related protein Rab-40C | Mus musculus (Mouse) | PR |
| P55258 | Rab8a | Ras-related protein Rab-8A | Mus musculus (Mouse) | PR |
| P35289 | Rab15 | Ras-related protein Rab-15 | Rattus norvegicus (Rat) | PR |
| P70550 | Rab8b | Ras-related protein Rab-8B | Rattus norvegicus (Rat) | PR |
| P35284 | Rab12 | Ras-related protein Rab-12 | Rattus norvegicus (Rat) | PR |
| P35281 | Rab10 | Ras-related protein Rab-10 | Rattus norvegicus (Rat) | PR |
| P35280 | Rab8a | Ras-related protein Rab-8A | Rattus norvegicus (Rat) | PR |
| O24466 | RABE1A | Ras-related protein RABE1a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SF91 | RABE1E | Ras-related protein RABE1e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LZD4 | RABE1D | Ras-related protein RABE1d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAKTYDYLFK | LLLIGDSGVG | KTCALFRFSE | DAFNATFIST | IGIDFKIRTI | ELDGKRIKLQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IWDTAGQERF | RTITTAYYRG | AMGIMLVYDI | TNEKSFENIR | NWVRNIEEHA | SPDVEKMILG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NKCDANDKRQ | VSREQGEKLA | ASFGIKFMET | SAKANINIEN | AFFTLARDIK | AKMDKKLEGN |
| 190 | 200 | ||||
| SPQGSNQGVK | ITPDQQKKSS | FFRCVLL |