Q5EAD0
Gene name |
BAIAP2 |
Protein name |
Brain-specific angiogenesis inhibitor 1-associated protein 2 |
Names |
BAI-associated protein 2, BAI1-associated protein 2 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:507837 |
EC number |
|
Protein Class |
|
Descriptions
(Annotation based on sequence homology with Q9UQB8)
BAIAP2 (IRSp53) is a SH3-containing protein that links membrane-bound small G-proteins to cytoplasmic effector proteins. The N-terminal autoinhibitory region interacts intramolecularly with the central region containing CRIB motif and Cdc42 binding domain. When GTP-loaded Cdc42 binds to the CRIB motif, it abrogates the autoinhibitory, intramolecular interaction and allows the SH3 domain to interact with BAIAP2 effector proteins such as Mena. When the N-terminal autoinhibitory fragment is co-expressed with BAIAP2, the fragment inhibits the function of BAIAP2.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
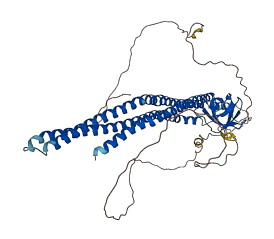
Autoinhibited structure

Activated structure

1 structures for Q5EAD0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5EAD0-F1 | Predicted | AlphaFoldDB |
151 variants for Q5EAD0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs439495013 | 2 | S>L | No | EVA | |
| rs458055016 | 2 | S>P | No | EVA | |
| rs442155045 | 6 | S>A | No | EVA | |
| rs442155045 | 6 | S>P | No | EVA | |
| rs475102910 | 7 | E>G | No | EVA | |
| rs456578642 | 8 | E>D | No | EVA | |
| rs444611710 | 9 | M>V | No | EVA | |
| rs470971145 | 10 | H>P | No | EVA | |
| rs452724177 | 11 | R>G | No | EVA | |
| rs465644505 | 12 | L>H | No | EVA | |
| rs465644505 | 12 | L>P | No | EVA | |
| rs453629247 | 13 | T>P | No | EVA | |
| rs468248259 | 14 | E>D | No | EVA | |
| rs435132854 | 14 | E>K | No | EVA | |
| rs449737803 | 15 | N>H | No | EVA | |
| rs464342475 | 15 | N>K | No | EVA | |
| rs482905547 | 15 | N>S | No | EVA | |
| rs478905046 | 16 | V>A | No | EVA | |
| rs445991055 | 16 | V>F | No | EVA | |
| rs478905046 | 16 | V>G | No | EVA | |
| rs481432417 | 17 | Y>* | No | EVA | |
| rs460578167 | 17 | Y>H | No | EVA | |
| rs442007072 | 17 | Y>S | No | EVA | |
| rs463023865 | 18 | K>R | No | EVA | |
| rs460651143 | 45 | V>G | No | EVA | |
| rs440815522 | 47 | Y>D | No | EVA | |
| rs461895017 | 54 | D>G | No | EVA | |
| rs476257784 | 58 | K>Q | No | EVA | |
| rs451535497 | 61 | E>A | No | EVA | |
| rs432919632 | 69 | S>T | No | EVA | |
| rs453926188 | 70 | K>Q | No | EVA | |
| rs437491174 | 74 | D>E | No | EVA | |
| rs470301581 | 77 | F>C | No | EVA | |
| rs468213930 | 98 | H>P | No | EVA | |
| rs450045551 | 115 | Y>S | No | EVA | |
| rs464530646 | 117 | S>R | No | EVA | |
| rs521541519 | 133 | A>V | No | EVA | |
| rs459394520 | 172 | Q>E | No | EVA | |
| rs461743547 | 174 | E>G | No | EVA | |
| rs476518482 | 175 | L>R | No | EVA | |
| rs443227215 | 175 | L>V | No | EVA | |
| rs451510089 | 177 | S>G | No | EVA | |
| rs432903828 | 177 | S>R | No | EVA | |
| rs472205631 | 178 | Y>S | No | EVA | |
| rs453824509 | 185 | T>P | No | EVA | |
| rs435673973 | 188 | T>P | No | EVA | |
| rs450044346 | 194 | F>L | No | EVA | |
| rs438006311 | 194 | F>L | No | EVA | |
| rs450044346 | 194 | F>V | No | EVA | |
| rs464826114 | 196 | F>S | No | EVA | |
| rs446364833 | 197 | L>M | No | EVA | |
| rs479354512 | 198 | V>A | No | EVA | |
| rs460728945 | 199 | E>G | No | EVA | |
| rs448737802 | 200 | K>T | No | EVA | |
| rs461842723 | 201 | Q>P | No | EVA | |
| rs461842723 | 201 | Q>R | No | EVA | |
| rs443180787 | 203 | A>P | No | EVA | |
| rs476213546 | 209 | A>P | No | EVA | |
| rs448750847 | 212 | H>L | No | EVA | |
| rs448750847 | 212 | H>P | No | EVA | |
| rs439458827 | 214 | K>* | No | EVA | |
| rs463462326 | 215 | G>A | No | EVA | |
| rs471392934 | 216 | K>R | No | EVA | |
| rs434668783 | 217 | E>G | No | EVA | |
| rs452877317 | 217 | E>K | No | EVA | |
| rs474137374 | 226 | W>* | No | EVA | |
| rs455495353 | 228 | Q>P | No | EVA | |
| rs473733323 | 230 | C>W | No | EVA | |
| rs436996344 | 232 | D>H | No | EVA | |
| rs470070918 | 237 | P>A | No | EVA | |
| rs445306599 | 238 | D>A | No | EVA | |
| rs433299145 | 240 | A>G | No | EVA | |
| rs465996065 | 242 | Q>P | No | EVA | |
| rs447621899 | 243 | L>M | No | EVA | |
| rs480927568 | 247 | M>R | No | EVA | |
| rs462443938 | 248 | G>D | No | EVA | |
| rs450228516 | 249 | N>T | No | EVA | |
| rs483215504 | 252 | G>A | No | EVA | |
| rs463400852 | 253 | S>P | No | EVA | |
| rs438514125 | 254 | I>T | No | EVA | |
| rs471532008 | 257 | S>T | No | EVA | |
| rs459327353 | 259 | L>F | No | EVA | |
| rs440816058 | 262 | S>P | No | EVA | |
| rs443124225 | 267 | V>F | No | EVA | |
| rs474093450 | 270 | D>A | No | EVA | |
| rs455562104 | 271 | P>T | No | EVA | |
| rs800794102 | 297 | A>T | No | EVA | |
| rs452832728 | 357 | T>N | No | EVA | |
| rs434322547 | 359 | N>H | No | EVA | |
| rs467130079 | 361 | T>P | No | EVA | |
| rs448702914 | 362 | L>R | No | EVA | |
| rs449555493 | 366 | S>R | No | EVA | |
| rs457831692 | 368 | M>I | No | EVA | |
| rs482432841 | 368 | M>K | No | EVA | |
| rs445901430 | 371 | G>R | No | EVA | |
| rs478994147 | 372 | L>M | No | EVA | |
| rs460286620 | 372 | L>R | No | EVA | |
| rs441834501 | 374 | R>P | No | EVA | |
| rs475074572 | 375 | N>I | No | EVA | |
| rs475074572 | 375 | N>S | No | EVA | |
| rs444400601 | 378 | M>V | No | EVA | |
| rs470981948 | 380 | V>G | No | EVA | |
| rs452570616 | 382 | A>P | No | EVA | |
| rs473757751 | 383 | I>M | No | EVA | |
| rs434263605 | 383 | I>V | No | EVA | |
| rs455018570 | 385 | S>P | No | EVA | |
| rs436602450 | 388 | A>S | No | EVA | |
| rs468062678 | 390 | D>E | No | EVA | |
| rs449694026 | 393 | T>P | No | EVA | |
| rs463979425 | 399 | E>G | No | EVA | |
| rs445656854 | 400 | G>C | No | EVA | |
| rs445656854 | 400 | G>S | No | EVA | |
| rs478926773 | 402 | L>P | No | EVA | |
| rs460476317 | 404 | T>P | No | EVA | |
| rs460476317 | 404 | T>S | No | EVA | |
| rs448146241 | 405 | L>P | No | EVA | |
| rs481216901 | 406 | L>V | No | EVA | |
| rs462797009 | 408 | P>T | No | EVA | |
| rs444537663 | 409 | E>Q | No | EVA | |
| rs458913079 | 414 | W>C | No | EVA | |
| rs440414488 | 419 | S>R | No | EVA | |
| rs473660752 | 423 | K>T | No | EVA | |
| rs473793205 | 427 | W>L | No | EVA | |
| rs455408557 | 428 | F>V | No | EVA | |
| rs437235667 | 429 | P>H | No | EVA | |
| rs451559495 | 431 | S>F | No | EVA | |
| rs466434758 | 433 | T>N | No | EVA | |
| rs433181406 | 433 | T>P | No | EVA | |
| rs480995322 | 434 | R>P | No | EVA | |
| rs468756588 | 436 | L>P | No | EVA | |
| rs481890043 | 437 | D>A | No | EVA | |
| rs463466242 | 437 | D>E | No | EVA | |
| rs450344722 | 437 | D>H | No | EVA | |
| rs438344693 | 439 | D>H | No | EVA | |
| rs459388869 | 442 | D>G | No | EVA | |
| rs459388869 | 442 | D>V | No | EVA | |
| rs441075851 | 444 | L>V | No | EVA | |
| rs473931911 | 445 | H>L | No | EVA | |
| rs455343500 | 445 | H>Q | No | EVA | |
| rs434181141 | 447 | S>R | No | EVA | |
| rs447297441 | 472 | Y>S | No | EVA | |
| rs467989099 | 487 | F>S | No | EVA | |
| rs449623120 | 490 | R>S | No | EVA | |
| rs482964940 | 496 | V>G | No | EVA | |
| rs464482506 | 497 | P>L | No | EVA | |
| rs460349982 | 499 | F>S | No | EVA | |
| rs478859124 | 499 | F>V | No | EVA | |
| rs442133039 | 501 | Q>R | No | EVA | |
| rs438065007 | 516 | V>G | No | EVA | |
| rs464663873 | 519 | A>G | No | EVA | |
| rs111023026 | 519 | A>S | No | EVA |
No associated diseases with Q5EAD0
14 regional properties for Q5EAD0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | B-box-type zinc finger | 94 - 135 | IPR000315 |
| domain | Zinc finger, RING-type | 16 - 61 | IPR001841 |
| domain | B30.2/SPRY domain | 292 - 486 | IPR001870 |
| domain | SPRY domain | 362 - 485 | IPR003877 |
| domain | Butyrophylin-like, SPRY domain | 308 - 325 | IPR003879-1 |
| domain | Butyrophylin-like, SPRY domain | 325 - 342 | IPR003879-2 |
| domain | Butyrophylin-like, SPRY domain | 347 - 371 | IPR003879-3 |
| domain | Butyrophylin-like, SPRY domain | 379 - 392 | IPR003879-4 |
| domain | Butyrophylin-like, SPRY domain | 422 - 446 | IPR003879-5 |
| domain | Butyrophylin-like, SPRY domain | 452 - 470 | IPR003879-6 |
| domain | SPRY-associated | 309 - 361 | IPR006574 |
| conserved_site | Zinc finger, RING-type, conserved site | 31 - 40 | IPR017907 |
| domain | Zinc finger, C3HC4 RING-type | 16 - 60 | IPR018957 |
| domain | TRIM10, RING-HC finger | 9 - 69 | IPR042784 |
Functions
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| cytoskeletal anchor activity | The binding activity of a protein that brings together a cytoskeletal protein (either a microtubule or actin filament, spindle pole body, or protein directly bound to them) and one or more other molecules, permitting them to function in a coordinated way. |
| proline-rich region binding | Binding to a proline-rich region, i.e. a region that contains a high proportion of proline residues, in a protein. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| actin crosslink formation | The process in which two or more actin filaments are connected together by proteins that act as crosslinks between the filaments. The crosslinked filaments may be on the same or differing axes. |
| actin filament bundle assembly | The assembly of actin filament bundles; actin filaments are on the same axis but may be oriented with the same or opposite polarities and may be packed with different levels of tightness. |
| plasma membrane organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the plasma membrane. |
| positive regulation of actin cytoskeleton reorganization | Any process that activates or increases the frequency, rate or extent of actin cytoskeleton reorganization. |
| positive regulation of actin filament polymerization | Any process that activates or increases the frequency, rate or extent of actin polymerization. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| response to bacterium | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a bacterium. |
3 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9UQB8 | BAIAP2 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | Homo sapiens (Human) | EV |
| Q8BKX1 | Baiap2 | BAR/IMD domain-containing adapter protein 2 | Mus musculus (Mouse) | SS |
| Q6GMN2 | Baiap2 | BAR/IMD domain-containing adapter protein 2 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLSRSEEMH | RLTENVYKTI | MEQFNPSLRN | FIAMGKNYEK | ALAGVTYAAK | GYFDALVKMG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ELASESQGSK | ELGDVLFQMA | EVHRQIQNQL | EEMLKSFHNE | LLTQLEQKVE | LDSRYLSAAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KKYQTEQRSK | GDALDKCQAE | LKKLRKKSQG | SKNPQKYSDK | ELQYIDAIGN | KQGELESYVS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DGYKTALTEE | RRRFCFLVEK | QCAVAKNSAA | YHSKGKELLA | QKLPLWQQAC | ADPNKIPDRA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VQLMQQMGNS | NGSILPSGLS | ASKSNLVISD | PIPGAKPLPV | PPELAPFVGR | LSAQENAPVM |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NGVSGPDSED | YNPWADRKAT | QPKSTSPPQS | QSKLSDSYSN | TLPVRKSVAP | KNSYATTENK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TLPRSSSMAA | GLERNGRMRV | KAIFSHAAGD | NSTLLSFKEG | DLITLLVPEA | RDGWHYGESE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KTKMRGWFPF | SYTRVLDNDG | GDRLHMSLQQ | GKSSSTGNLL | DKEDLALPPP | DYGTSSRAFP |
| 490 | 500 | 510 | 520 | ||
| TQTAGAFKQR | PYSVAVPAFS | QGLDDYGARA | VSSADVEVAR | F |