Q5E9X2
Gene name |
MAP2K6 |
Protein name |
Dual specificity mitogen-activated protein kinase kinase 6 |
Names |
MAP kinase kinase 6, MAPKK 6, MAPK/ERK kinase 6, MEK 6 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:286883 |
EC number |
2.7.12.2: Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
Protein Class |
|
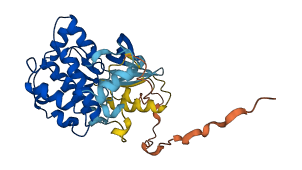
Descriptions
(Annotation based on sequence homology with P52564). MAP2K6 is Dual specificity protein kinase which acts as an essential component of the MAP kinase signal transduction pathway. The mechanism of autoinhibition of MAP2K6 differents from those observed for MAP2K1/4 regulation. The activation loop consisting of 3 short α-helices (AH1, AH2, AH3), which are significant in forming the autoinhibition state. AH1, AH2 and AH3 stabilizes the MAP2K6 in the inactive conformation by several interactions between themselves and main body of kinase domain. The phospho-AH2 presumably releases the molecular break and triggers a conformational transitions to the flexible states.
Autoinhibitory domains (AIDs)
Accessory elements
196-218 (Activation loop from InterPro)
Target domain |
53-314 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q5E9X2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5E9X2-F1 | Predicted | AlphaFoldDB |
41 variants for Q5E9X2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs470672358 | 12 | G>A | No | Ensembl | |
| rs463637168 | 48 | V>G | No | Ensembl | |
| rs441854262 | 52 | D>G | No | Ensembl | |
| rs474782667 | 56 | I>T | No | Ensembl | |
| rs483195700 | 58 | E>* | No | Ensembl | |
| rs451609830 | 64 | Y>S | No | Ensembl | |
| rs469681892 | 66 | V>A | No | Ensembl | |
| rs457460103 | 67 | V>G | No | Ensembl | |
| rs435779822 | 70 | M>R | No | Ensembl | |
| rs446544195 | 74 | P>A | No | Ensembl | |
| rs446544195 | 74 | P>S | No | Ensembl | |
| rs464214714 | 76 | E>D | No | Ensembl | |
| rs479324672 | 76 | E>G | No | Ensembl | |
| rs445611134 | 78 | I>N | No | Ensembl | |
| rs445611134 | 78 | I>S | No | Ensembl | |
| rs481831884 | 79 | M>I | No | Ensembl | |
| rs443493724 | 81 | V>G | No | Ensembl | |
| rs448468354 | 83 | R>P | No | Ensembl | |
| rs481265427 | 84 | I>S | No | Ensembl | |
| rs441148475 | 92 | E>Q | No | Ensembl | |
| rs459152360 | 105 | R>S | No | Ensembl | |
| rs468896996 | 135 | S>A | No | Ensembl | |
| rs1117023645 | 146 | K>I | No | Ensembl | |
| rs446103927 | 179 | D>E | No | Ensembl | |
| rs482067516 | 180 | V>G | No | Ensembl | |
| rs41932221 | 246 | T>S | No | Ensembl | |
| rs481996653 | 249 | E>G | No | Ensembl | |
| rs445896215 | 249 | E>Q | No | Ensembl | |
| rs1116681871 | 251 | A>T | No | Ensembl | |
| rs444518550 | 258 | D>E | No | Ensembl | |
| rs462158540 | 262 | T>A | No | Ensembl | |
| rs440368419 | 262 | T>I | No | Ensembl | |
| rs462158540 | 262 | T>P | No | Ensembl | |
| rs451877550 | 270 | V>G | No | Ensembl | |
| rs439810197 | 273 | E>A | No | Ensembl | |
| rs475801578 | 278 | L>P | No | Ensembl | |
| rs457426189 | 290 | F>C | No | Ensembl | |
| rs435276031 | 291 | T>P | No | Ensembl | |
| rs437033549 | 329 | K>R | No | Ensembl | |
| rs469978254 | 332 | L>R | No | Ensembl | |
| rs448221867 | 333 | G>A | No | Ensembl |
No associated diseases with Q5E9X2
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.12.2 | Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| MAP kinase kinase activity | Catalysis of the concomitant phosphorylation of threonine (T) and tyrosine (Y) residues in a Thr-Glu-Tyr (TEY) thiolester sequence in a MAP kinase (MAPK) substrate. |
| phosphatase activator activity | Binds to and increases the activity of a phosphatase, an enzyme which catalyzes of the removal of a phosphate group from a substrate molecule. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activator activity | Binds to and increases the activity of a protein serine/threonine kinase. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| bone development | The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components. |
| cardiac muscle contraction | Muscle contraction of cardiac muscle tissue. |
| negative regulation of cold-induced thermogenesis | Any process that stops, prevents, or reduces the rate of cold-induced thermogenesis. |
| osteoblast differentiation | The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, a mesodermal or neural crest cell that gives rise to bone. |
| positive regulation of MAP kinase activity | Any process that activates or increases the frequency, rate or extent of MAP kinase activity. |
| stress-activated MAPK cascade | The series of molecular signals in which a stress-activated MAP kinase cascade relays a signal; MAP kinase cascades involve at least three protein kinase activities and culminate in the phosphorylation and activation of a MAP kinase. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P46734 | MAP2K3 | Dual specificity mitogen-activated protein kinase kinase 3 | Homo sapiens (Human) | SS |
| P52564 | MAP2K6 | Dual specificity mitogen-activated protein kinase kinase 6 | Homo sapiens (Human) | EV |
| O14733 | MAP2K7 | Dual specificity mitogen-activated protein kinase kinase 7 | Homo sapiens (Human) | PR |
| P45985 | MAP2K4 | Dual specificity mitogen-activated protein kinase kinase 4 | Homo sapiens (Human) | EV |
| O09110 | Map2k3 | Dual specificity mitogen-activated protein kinase kinase 3 | Mus musculus (Mouse) | PR |
| P70236 | Map2k6 | Dual specificity mitogen-activated protein kinase kinase 6 | Mus musculus (Mouse) | EV |
| Q8CE90 | Map2k7 | Dual specificity mitogen-activated protein kinase kinase 7 | Mus musculus (Mouse) | PR |
| P47809 | Map2k4 | Dual specificity mitogen-activated protein kinase kinase 4 | Mus musculus (Mouse) | EV |
| G5EDF7 | sek-1 | Dual specificity mitogen-activated protein kinase kinase sek-1 | Caenorhabditis elegans | PR |
| G5EDT6 | jkk-1 | Dual specificity mitogen-activated protein kinase kinase jkk-1 | Caenorhabditis elegans | PR |
| Q21307 | mek-1 | Dual specificity mitogen-activated protein kinase kinase mek-1 | Caenorhabditis elegans | PR |
| Q9DGE0 | map2k6 | Dual specificity mitogen-activated protein kinase kinase 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSQSKGKKRN | PGLKIPKEAF | EQPQTSSTPP | RDLDSKACIS | IGNQNFEVKA | DDLEPIVELG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RGAYGVVEKM | RHVPSEQIMA | VKRIRATVNS | QEQKRLLMDL | DISMRTVDCP | FTVTFYGALF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| REGDVWICME | LMDTSLDKFY | KQVIDKGQTI | PEDILGKIAV | SIVKALEHLH | SKLSVIHRDV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KPSNVLINAL | GQVKMCDFGI | SGYLVDSVAK | TIDAGCKPYM | APERINPELN | QKGYSVKSDI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| WSLGITMIEL | AILRFPYDSW | GTPFQQLKQV | VEEPSPQLPA | DKFSEEFVDF | TSQCLKKNSK |
| 310 | 320 | 330 | |||
| ERPTYPELMQ | HPFFTLHESK | ATDVASFVKS | ILGD |