Q5E9F8
Gene name |
H3-3B |
Protein name |
Histone H3.3 |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:617051 |
EC number |
|
Protein Class |
|
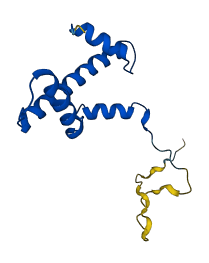
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q5E9F8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5E9F8-F1 | Predicted | AlphaFoldDB |
66 variants for Q5E9F8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs452542277 | 7 | T>P | No | EVA | |
| rs480255111 | 11 | S>T | No | EVA | |
| rs434683586 | 15 | K>E | No | EVA | |
| rs434683586 | 15 | K>Q | No | EVA | |
| rs445088683 | 16 | A>E | No | EVA | |
| rs433128548 | 17 | P>A | No | EVA | |
| rs456659256 | 19 | K>R | No | EVA | |
| rs476690688 | 20 | Q>K | No | EVA | |
| rs440092262 | 21 | L>I | No | EVA | |
| rs455864130 | 23 | T>N | No | EVA | |
| rs435831326 | 23 | T>P | No | EVA | |
| rs472796536 | 24 | K>N | No | EVA | |
| rs441506027 | 26 | A>G | No | EVA | |
| rs458247402 | 29 | S>N | No | EVA | |
| rs471237874 | 30 | A>D | No | EVA | |
| rs443168430 | 33 | T>P | No | EVA | |
| rs482598027 | 36 | V>A | No | EVA | |
| rs482598027 | 36 | V>G | No | EVA | |
| rs480118057 | 37 | K>E | No | EVA | |
| rs448920567 | 39 | P>T | No | EVA | |
| rs473787411 | 40 | H>N | No | EVA | |
| rs445116318 | 43 | R>T | No | EVA | |
| rs475626959 | 45 | G>R | No | EVA | |
| rs461245805 | 51 | E>V | No | EVA | |
| rs481360314 | 52 | I>T | No | EVA | |
| rs459757763 | 55 | Y>S | No | EVA | |
| rs479582219 | 59 | T>A | No | EVA | |
| rs458859525 | 63 | I>L | No | EVA | |
| rs482261390 | 64 | R>L | No | EVA | |
| rs467892670 | 65 | K>E | No | EVA | |
| rs436696316 | 65 | K>N | No | EVA | |
| rs447064677 | 66 | L>R | No | EVA | |
| rs468142895 | 68 | F>Y | No | EVA | |
| rs466604383 | 72 | V>L | No | EVA | |
| rs468781201 | 74 | E>A | No | EVA | |
| rs431929574 | 82 | D>A | No | EVA | |
| rs436068849 | 94 | Q>R | No | EVA | |
| rs482813892 | 95 | E>A | No | EVA | |
| rs448034930 | 96 | A>D | No | EVA | |
| rs448034930 | 96 | A>V | No | EVA | |
| rs457123894 | 97 | S>R | No | EVA | |
| rs468160218 | 98 | E>A | No | EVA | |
| rs433826898 | 98 | E>D | No | EVA | |
| rs453881626 | 99 | A>S | No | EVA | |
| rs464336900 | 100 | Y>D | No | EVA | |
| rs432934256 | 100 | Y>S | No | EVA | |
| rs456205604 | 102 | V>G | No | EVA | |
| rs444509468 | 104 | L>F | No | EVA | |
| rs433013230 | 108 | T>P | No | EVA | |
| rs453094978 | 112 | A>D | No | EVA | |
| rs473460395 | 114 | H>N | No | EVA | |
| rs475013148 | 120 | I>N | No | EVA | |
| rs468842166 | 122 | P>T | No | EVA | |
| rs442142948 | 125 | I>T | No | EVA | |
| rs455685167 | 126 | Q>R | No | EVA | |
| rs460832090 | 128 | A>P | No | EVA | |
| rs444743893 | 129 | R>C | No | EVA | |
| rs477730765 | 129 | R>P | No | EVA | |
| rs458404970 | 129 | R>P | No | EVA | |
| rs472006061 | 130 | R>P | No | EVA | |
| rs439809124 | 131 | I>V | No | EVA | |
| rs448094754 | 133 | G>R | No | EVA | |
| rs479273387 | 134 | E>K | No | EVA | |
| rs478640462 | 135 | R>G | No | EVA | |
| rs448132658 | 135 | R>H | No | EVA | |
| rs448132658 | 135 | R>L | No | EVA |
No associated diseases with Q5E9F8
1 regional properties for Q5E9F8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Histone H2A/H2B/H3 | 1 - 132 | IPR007125 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| Barr body | A structure found in a female mammalian cell containing an unpaired X chromosome that has become densely heterochromatic, silenced and localized at the nuclear periphery. |
| chromosome, telomeric region | The end of a linear chromosome, required for the integrity and maintenance of the end. A chromosome telomere usually includes a region of telomerase-encoded repeats the length of which rarely exceeds 20 bp each and that permits the formation of a telomeric loop (T-loop). The telomeric repeat region is usually preceded by a sub-telomeric region that is gene-poor but rich in repetitive elements. Some telomeres only consist of the latter part (for eg. D. melanogaster telomeres). |
| inner kinetochore | The region of a kinetochore closest to centromeric DNA; in mammals the CREST antigens (CENP proteins) are found in this layer; this layer may help define underlying centromeric chromatin structure and position of the kinetochore on the chromosome. |
| nucleosome | A complex comprised of DNA wound around a multisubunit core and associated proteins, which forms the primary packing unit of DNA into higher order structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| protein heterodimerization activity | Binding to a nonidentical protein to form a heterodimer. |
| structural constituent of chromatin | The action of a molecule that contributes to the structural integrity of chromatin. |
14 GO annotations of biological process
| Name | Definition |
|---|---|
| cell population proliferation | The multiplication or reproduction of cells, resulting in the expansion of a cell population. |
| embryo implantation | Attachment of the blastocyst to the uterine lining. |
| male gonad development | The process whose specific outcome is the progression of the male gonad over time, from its formation to the mature structure. |
| multicellular organism growth | The increase in size or mass of an entire multicellular organism, as opposed to cell growth. |
| muscle cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a muscle cell. |
| negative regulation of chromosome condensation | Any process that stops, prevents or reduces the frequency, rate or extent of chromosome condensation. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| oocyte maturation | A developmental process, independent of morphogenetic (shape) change, that is required for an oocyte to attain its fully functional state. Oocyte maturation commences after reinitiation of meiosis commonly starting with germinal vesicle breakdown, and continues up to the second meiotic arrest prior to fertilization. |
| osteoblast differentiation | The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, a mesodermal or neural crest cell that gives rise to bone. |
| pericentric heterochromatin assembly | The compaction of chromatin located adjacent to the CENP-A rich centromere 'central core' and characterized by methylation of histone H3K9, into heterochromatin, resulting in the repression of transcription at pericentric DNA. |
| regulation of centromere complex assembly | Any process that modulates the rate, frequency, or extent of centromere complex assembly, the aggregation, arrangement and bonding together of proteins and centromeric DNA molecules to form a centromeric protein-DNA complex. |
| single fertilization | The union of male and female gametes to form a zygote. |
| spermatid development | The process whose specific outcome is the progression of a spermatid over time, from its formation to the mature structure. |
| subtelomeric heterochromatin assembly | The compaction of chromatin into heterochromatin at the subtelomeric region. |
38 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P68428 | Histone H3.2 | Triticum aestivum (Wheat) | PR | |
| Q5E9F8 | H3-3B | Histone H3.3 | Bos taurus (Bovine) | PR |
| P68432 | Histone H3.1 | Bos taurus (Bovine) | PR | |
| P84227 | Histone H3.2 | Bos taurus (Bovine) | PR | |
| P84229 | H3-VIII | Histone H3.2 | Gallus gallus (Chicken) | PR |
| P84247 | H3-X | Histone H3.3 | Gallus gallus (Chicken) | PR |
| Q71V89 | HIS3 | Histone H3.3 | Gossypium hirsutum (Upland cotton) (Gossypium mexicanum) | PR |
| P02299 | His3 | Histone H3 | Drosophila melanogaster (Fruit fly) | PR |
| Q71H73 | Histone H3.3 | Vitis vinifera (Grape) | PR | |
| Q71DI3 | H3C13 | Histone H3.2 | Homo sapiens (Human) | PR |
| P68431 | H3C12 | Histone H3.1 | Homo sapiens (Human) | PR |
| Q16695 | H3-4 | Histone H3.1t | Homo sapiens (Human) | PR |
| P84243 | H3-3B | Histone H3.3 | Homo sapiens (Human) | PR |
| P69246 | H3C4 | Histone H3.2 | Zea mays (Maize) | PR |
| P84228 | H3c15 | Histone H3.2 | Mus musculus (Mouse) | PR |
| P68433 | H3c11 | Histone H3.1 | Mus musculus (Mouse) | PR |
| P84244 | H3-3b | Histone H3.3 | Mus musculus (Mouse) | PR |
| Q71LE2 | H3-3A | Histone H3.3 | Sus scrofa (Pig) | PR |
| O35799 | Hfe | Hereditary hemochromatosis protein homolog | Rattus norvegicus (Rat) | PR |
| Q6LED0 | Histone H3.1 | Rattus norvegicus (Rat) | PR | |
| P84245 | H3-3b | Histone H3.3 | Rattus norvegicus (Rat) | PR |
| Q0JCT1 | H3 | Histone H3.3 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAD9 | H3R-21 | Histone H3.2 | Oryza sativa subsp japonica (Rice) | PR |
| P08898 | his-2 | Histone H3 | Caenorhabditis elegans | PR |
| Q10453 | his-71 | Histone H3.3 type 1 | Caenorhabditis elegans | PR |
| Q27490 | his-70 | Histone H3.3-like type 1 | Caenorhabditis elegans | PR |
| Q27532 | his-74 | Histone H3.3-like type 2 | Caenorhabditis elegans | PR |
| Q9U281 | his-72 | Histone H3.3 type 2 | Caenorhabditis elegans | PR |
| Q9FKQ3 | At5g65350 | Histone H3-like 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P59226 | HTR2 | Histone H3.1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FX60 | At1g13370 | Histone H3-like 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FXI7 | MGH3 | Histone H3-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LR02 | At1g75600 | Histone H3-like 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P59169 | HTR4 | Histone H3.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q28D37 | TGas081o10.1 | Histone H3.2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q6P823 | TGas113e22.1 | Histone H3.3 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q4QRF4 | zgc:113984; | Histone H3.2 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q6PI20 | h3f3a | Histone H3.3 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARTKQTARK | STGGKAPRKQ | LATKAARKSA | PSTGGVKKPH | RYRPGTVALR | EIRRYQKSTE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LLIRKLPFQR | LVREIAQDFK | TDLRFQSAAI | GALQEASEAY | LVGLFEDTNL | CAIHAKRVTI |
| 130 | |||||
| MPKDIQLARR | IRGERA |