Q5DU57
Gene name |
Spata13 |
Protein name |
Spermatogenesis-associated protein 13 |
Names |
APC-stimulated guanine nucleotide exchange factor 2, Asef2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:219140 |
EC number |
|
Protein Class |
|
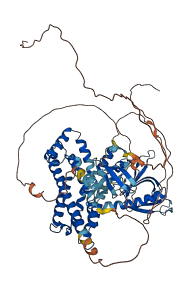
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
561-652 (C-terminal tail) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Hamann MJ et al. (2007) "Asef2 functions as a Cdc42 exchange factor and is stimulated by the release of an autoinhibitory module from a concealed C-terminal activation element", Molecular and cellular biology, 27, 1380-93
- Murayama K et al. (2007) "Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism", The Journal of biological chemistry, 282, 4238-4242
- Mitin N et al. (2007) "Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression", Nature structural & molecular biology, 14, 814-23
- Ravala SK et al. (2020) "The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity and contributes to membrane binding", The Journal of biological chemistry, 295, 12635-12647
Autoinhibited structure

Activated structure

1 structures for Q5DU57
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q5DU57-F1 | Predicted | AlphaFoldDB |
No variants for Q5DU57
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q5DU57 | |||||
No associated diseases with Q5DU57
13 regional properties for Q5DU57
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Receptor L-domain | 356 - 463 | IPR000494-1 |
| domain | Receptor L-domain | 663 - 777 | IPR000494-2 |
| domain | Protein kinase domain | 1371 - 1659 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 1375 - 1649 | IPR001245 |
| conserved_site | Tyrosine-protein kinase, receptor class II, conserved site | 1543 - 1551 | IPR002011 |
| domain | Fibronectin type III | 805 - 910 | IPR003961-1 |
| domain | Fibronectin type III | 929 - 1175 | IPR003961-2 |
| domain | Fibronectin type III | 1208 - 1305 | IPR003961-3 |
| domain | Furin-like cysteine-rich domain | 510 - 648 | IPR006211 |
| repeat | Furin-like repeat | 542 - 592 | IPR006212 |
| active_site | Tyrosine-protein kinase, active site | 1515 - 1527 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 1377 - 1405 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 1371 - 1650 | IPR020635 |
Functions
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| guanyl-nucleotide exchange factor activity | Stimulates the exchange of GDP to GTP on a signaling GTPase, changing its conformation to its active form. Guanine nucleotide exchange factors (GEFs) act by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP), which is more abundant in the cell under normal cellular physiological conditions. |
| identical protein binding | Binding to an identical protein or proteins. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| filopodium assembly | The assembly of a filopodium, a thin, stiff protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal growth cone. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| lamellipodium assembly | Formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| regulation of cell migration | Any process that modulates the frequency, rate or extent of cell migration. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q58DL7 | ARHGEF9 | Rho guanine nucleotide exchange factor 9 | Bos taurus (Bovine) | SS |
| O43307 | ARHGEF9 | Rho guanine nucleotide exchange factor 9 | Homo sapiens (Human) | SS |
| Q70Z35 | PREX2 | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 protein | Homo sapiens (Human) | SS |
| Q6XZF7 | DNMBP | Dynamin-binding protein | Homo sapiens (Human) | PR |
| Q9NR80 | ARHGEF4 | Rho guanine nucleotide exchange factor 4 | Homo sapiens (Human) | EV |
| A1IGU5 | ARHGEF37 | Rho guanine nucleotide exchange factor 37 | Homo sapiens (Human) | PR |
| Q8TCU6 | PREX1 | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | Homo sapiens (Human) | EV |
| Q96N96 | SPATA13 | Spermatogenesis-associated protein 13 | Homo sapiens (Human) | EV |
| Q3LAC4 | Prex2 | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 protein | Mus musculus (Mouse) | SS |
| Q69ZK0 | Prex1 | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | Mus musculus (Mouse) | SS |
| Q3UTH8 | Arhgef9 | Rho guanine nucleotide exchange factor 9 | Mus musculus (Mouse) | SS |
| Q7TNR9 | Arhgef4 | Rho guanine nucleotide exchange factor 4 | Mus musculus (Mouse) | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MHPASVTTTS | QDPCAPSGSC | RGGRRRRPIS | VIGGVSFYGN | TQVEDVENLL | VQPAARPPVP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AHQVPPYKAV | SARLRPFTFS | QSTPIGLDRV | GRRRQMKTSN | VSSDGGAESS | ALVDDNGSEE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DFSYEELCQA | NPRYLQPGGE | QLAINELISD | GSVVCAEALW | DHVTMDDQEL | GFKAGDVIQV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LEASNKDWWW | GRNEDKEAWF | PASFVRLRVN | QEELPENCSS | SHGEEQDEDT | SKARHKHPES |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QQQMRTNVIQ | EIMNTERVYI | KHLKDICEGY | IRQCRKHTGM | FTVAQLATIF | GNIEDIYKFQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RKFLKDLEKQ | YNKEEPHLSE | IGSCFLEHQE | GFAIYSEYCN | NHPGACVELS | NLMKHSKYRH |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FFEACRLLQQ | MIDIALDGFL | LTPVQKICKY | PLQLAELLKY | TTQEHGDYNN | IKAAYEAMKN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VACLINERKR | KLESIDKIAR | WQVSIVGWEG | LDILDRSSEL | IHSGELTKIT | RQGKSQQRIF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| FLFDHQLVSC | KKDLLRRDML | YYKGRMDMDE | VELVDVEDGR | DKDWSLSLRN | AFKLVSKATD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EVHLFCARKQ | EDKARWLQAY | ADERRRVQED | QQMGMEIPEN | QKKLAMLNAQ | KAGHGKSKGY |
| 610 | 620 | 630 | 640 | 650 | |
| NSCPVAPPHQ | SLPPLHQRHI | TVPTSIPQQQ | VFALAEPKRK | PSIFWHTFHK | LTPFRK |