Q58CP9
Gene name |
STIM1 |
Protein name |
Stromal interaction molecule 1 |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:534816 |
EC number |
|
Protein Class |
|
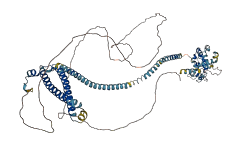
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
339-439 (SOAR domain) |
Relief mechanism |
Others |
Assay |
|
Target domain |
339-439 (SOAR domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Zheng L et al. (2011) "Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry", Proceedings of the National Academy of Sciences of the United States of America, 108, 1337-42
- Korzeniowski MK et al. (2010) "Activation of STIM1-Orai1 involves an intramolecular switching mechanism", Science signaling, 3, ra82
Autoinhibited structure

Activated structure

1 structures for Q58CP9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q58CP9-F1 | Predicted | AlphaFoldDB |
No variants for Q58CP9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q58CP9 | |||||
No associated diseases with Q58CP9
Functions
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| integral component of endoplasmic reticulum membrane | The component of the endoplasmic reticulum membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| plasma membrane raft | A membrane raft that is part of the plasma membrane. |
| sarcoplasmic reticulum membrane | The lipid bilayer surrounding the sarcoplasmic reticulum. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium channel regulator activity | Modulates the activity of a calcium channel. |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| microtubule plus-end binding | Binding to the plus end of a microtubule. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of store-operated calcium channel activity | A process that initiates the activity of an inactive store-operated calcium channel. |
| cellular calcium ion homeostasis | Any process involved in the maintenance of an internal steady state of calcium ions at the level of a cell. |
| detection of calcium ion | The series of events in which a calcium ion stimulus is received by a cell and converted into a molecular signal. |
| enamel mineralization | The process in which calcium salts, mainly carbonated hydroxyapatite, are deposited in tooth enamel. |
| positive regulation of adenylate cyclase activity | Any process that activates or increases the frequency, rate or extent of adenylate cyclase activity. |
| regulation of calcium ion transport | Any process that modulates the frequency, rate or extent of the directed movement of calcium ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of store-operated calcium entry | Any process that modulates the frequency, rate or extent of store-operated calcium entry. |
| store-operated calcium entry | A calcium ion entry mechanism in the plasma membrane activated by the depletion of calcium ion from the internal calcium ion store in the endoplasmic reticulum. |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P83094 | Stim | Stromal interaction molecule homolog | Drosophila melanogaster (Fruit fly) | SS |
| Q13586 | STIM1 | Stromal interaction molecule 1 | Homo sapiens (Human) | EV |
| P70302 | Stim1 | Stromal interaction molecule 1 | Mus musculus (Mouse) | SS |
| P84903 | Stim1 | Stromal interaction molecule 1 | Rattus norvegicus (Rat) | SS |
| G5EF60 | stim-1 | Stromal interaction molecule 1 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDVCARLALW | LLWGLLLHHG | QSLSQSHSEK | ATGSGANSEE | STAAEFCRID | KPLCHSEDEK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LSFDAVRSIH | KLMDDDANGD | VDVEESDEFL | REDLNYHDPT | VKHSTFHGED | KLISVEDLWK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AWKSSEVYNW | TVDEVVQWLI | TYVELPQYEE | TFRKLQLSGH | AMPRLAVTNT | TMTGTVLKMT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DRSHRQKLQL | KALDTVLFGP | PLLTRHNHLK | DFMLVVSIVI | GVGGCWFAYI | QNRYSKEHMK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KMMKDLEGLH | RAEQSLHDLQ | ERLHKAQEEH | RTVEVEKVHL | EKKLRDEINL | AKQEAQRLKE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LREGTENERS | RQKYAEEELE | QVREALRKAE | KELESHSSWY | APEALQKWLQ | LTHEVEVQYY |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NIKKQNAEKQ | LLVAKEGAEK | IKKKRNTLFG | TFHVAHSSSL | DDVDHKILTA | KQALSEVTAA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LRERLHRWQQ | IEILCGFQIV | NNPGIHSLVA | ALNIDPSWMG | STRPNPAHFI | MTDDVDDMDE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EIVSPLSMQS | PSLQSSVRQR | LTEPQHGLGS | QRDLTHSDSE | SSLHMSDRQR | LAPKPPQMIR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AADEALSAMT | SNGSHRLIEG | AHPGSLVEKL | PDSPALAKKA | LLALNHGLDK | AHSLMELSSP |
| 610 | 620 | 630 | 640 | 650 | 660 |
| ALPSGSPHLD | SSRSHSPSPP | DPDTPSPAGD | SRALQASRNT | RIPHLAGKKA | AAEEDNGSIG |
| 670 | 680 | ||||
| EETDSSPGRK | KFPLKIFKKP | LKK |