Q56X76
Gene name |
RH39 (At4g09730, F17A8.80) |
Protein name |
DEAD-box ATP-dependent RNA helicase 39 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT4G09730 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
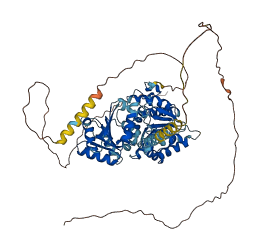
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q56X76
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q56X76-F1 | Predicted | AlphaFoldDB |
61 variants for Q56X76
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_4_6136361_C_T | 10 | S>F | No | 1000Genomes | |
| ENSVATH11393460 | 23 | I>T | No | 1000Genomes | |
|
ENSVATH02765336 tmp_4_6136441_T_C |
37 | F>L | No | 1000Genomes | |
| tmp_4_6136446_C_A | 38 | H>Q | No | 1000Genomes | |
| ENSVATH06593375 | 39 | H>R | No | 1000Genomes | |
| tmp_4_6136481_C_T | 50 | T>I | No | 1000Genomes | |
| tmp_4_6136487_C_T | 52 | S>F | No | 1000Genomes | |
| ENSVATH11393462 | 58 | T>I | No | 1000Genomes | |
| ENSVATH02765338 | 66 | L>M | No | 1000Genomes | |
| tmp_4_6136599_G_C | 89 | Q>H | No | 1000Genomes | |
| tmp_4_6136601_G_A | 90 | R>K | No | 1000Genomes | |
| ENSVATH11393463 | 90 | R>S | No | 1000Genomes | |
| ENSVATH02765342 | 98 | E>D | No | 1000Genomes | |
| ENSVATH06593377 | 101 | S>G | No | 1000Genomes | |
| tmp_4_6136649_A_T | 106 | K>M | No | 1000Genomes | |
| tmp_4_6136672_T_C | 114 | F>L | No | 1000Genomes | |
| ENSVATH06593378 | 121 | E>A | No | 1000Genomes | |
| ENSVATH02765344 | 128 | Q>R | No | 1000Genomes | |
| tmp_4_6136761_A_G | 143 | I>M | No | 1000Genomes | |
| tmp_4_6136822_C_T | 164 | L>F | No | 1000Genomes | |
| tmp_4_6136840_A_T | 170 | I>F | No | 1000Genomes | |
| tmp_4_6137478_G_A | 209 | A>T | No | 1000Genomes | |
| tmp_4_6137536_G_A | 228 | R>Q | No | 1000Genomes | |
| ENSVATH02765374 | 254 | I>V | No | 1000Genomes | |
| tmp_4_6137643_A_G | 264 | I>V | No | 1000Genomes | |
| ENSVATH06593413 | 265 | A>S | No | 1000Genomes | |
| ENSVATH00483601 | 286 | K>R | No | 1000Genomes | |
| ENSVATH06593421 | 289 | A>T | No | 1000Genomes | |
| ENSVATH11393513 | 295 | A>T | No | 1000Genomes | |
| tmp_4_6137833_C_T | 295 | A>V | No | 1000Genomes | |
| tmp_4_6137844_A_T | 299 | N>Y | No | 1000Genomes | |
| ENSVATH11393514 | 300 | D>G | No | 1000Genomes | |
| ENSVATH11393535 | 306 | V>I | No | 1000Genomes | |
| ENSVATH14112628 | 309 | T>I | No | 1000Genomes | |
| tmp_4_6138052_G_T | 323 | E>D | No | 1000Genomes | |
| ENSVATH06593431 | 328 | E>V | No | 1000Genomes | |
| ENSVATH14112655 | 345 | D>V | No | 1000Genomes | |
| tmp_4_6138346_G_A | 371 | S>N | No | 1000Genomes | |
| tmp_4_6138421_C_A | 396 | S>Y | No | 1000Genomes | |
| tmp_4_6138605_A_T | 425 | T>S | No | 1000Genomes | |
| ENSVATH11393542 | 476 | S>G | No | 1000Genomes | |
| tmp_4_6139098_G_A | 485 | R>Q | No | 1000Genomes | |
| ENSVATH11393543 | 508 | A>D | No | 1000Genomes | |
| ENSVATH02765394 | 511 | T>S | No | 1000Genomes | |
| ENSVATH02765395 | 512 | H>Q | No | 1000Genomes | |
| tmp_4_6139210_G_C | 522 | K>N | No | 1000Genomes | |
| tmp_4_6139211_C_G | 523 | Q>E | No | 1000Genomes | |
| ENSVATH06593475 | 531 | R>Q | No | 1000Genomes | |
| ENSVATH06593476 | 533 | S>T | No | 1000Genomes | |
| ENSVATH02765396 | 539 | S>T | No | 1000Genomes | |
| tmp_4_6139269_C_G | 542 | P>R | No | 1000Genomes | |
| ENSVATH02765398 | 542 | P>S | No | 1000Genomes | |
| ENSVATH11393544 | 546 | S>P | No | 1000Genomes | |
| ENSVATH00483614 | 547 | T>I | No | 1000Genomes | |
| ENSVATH00483614 | 547 | T>R | No | 1000Genomes | |
| ENSVATH06593478 | 552 | P>L | No | 1000Genomes | |
| ENSVATH02765400 | 565 | R>K | No | 1000Genomes | |
| tmp_4_6139395_C_T | 584 | T>M | No | 1000Genomes | |
| tmp_4_6139425_G_C | 594 | G>A | No | 1000Genomes | |
| tmp_4_6139427_A_G | 595 | K>E | No | 1000Genomes | |
| ENSVATH02765401 | 607 | S>R | No | 1000Genomes |
No associated diseases with Q56X76
4 regional properties for Q56X76
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| chloroplast | A chlorophyll-containing plastid with thylakoids organized into grana and frets, or stroma thylakoids, and embedded in a stroma. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| large ribosomal subunit rRNA binding | Binding to large ribosomal subunit RNA (LSU rRNA), a constituent of the large ribosomal subunit. In S. cerevisiae, this is the 25S rRNA. |
| mRNA binding | Binding to messenger RNA (mRNA), an intermediate molecule between DNA and protein. mRNA includes UTR and coding sequences, but does not contain introns. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| chloroplast rRNA processing | Any rRNA processing that takes place in chloroplast. |
| mitochondrial large ribosomal subunit assembly | The aggregation, arrangement and bonding together of a set of components to form a mitochondrial large ribosomal subunit. |
| ribulose bisphosphate carboxylase complex assembly | The aggregation, arrangement and bonding together of a set of components to form a ribulose bisphosphate carboxylase complex. |
28 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2NL08 | DDX55 | ATP-dependent RNA helicase DDX55 | Bos taurus (Bovine) | PR |
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | Homo sapiens (Human) | PR |
| Q8NHQ9 | DDX55 | ATP-dependent RNA helicase DDX55 | Homo sapiens (Human) | PR |
| Q9NUL7 | DDX28 | Probable ATP-dependent RNA helicase DDX28 | Homo sapiens (Human) | PR |
| Q99MJ9 | Ddx50 | ATP-dependent RNA helicase DDX50 | Mus musculus (Mouse) | PR |
| Q61656 | Ddx5 | Probable ATP-dependent RNA helicase DDX5 | Mus musculus (Mouse) | PR |
| Q501J6 | Ddx17 | Probable ATP-dependent RNA helicase DDX17 | Mus musculus (Mouse) | PR |
| Q6ZPL9 | Ddx55 | ATP-dependent RNA helicase DDX55 | Mus musculus (Mouse) | PR |
| Q9CWT6 | Ddx28 | Probable ATP-dependent RNA helicase DDX28 | Mus musculus (Mouse) | PR |
| Q5QMN3 | Os01g0197200 | DEAD-box ATP-dependent RNA helicase 20 | Oryza sativa subsp japonica (Rice) | PR |
| Q5N7W4 | Os01g0911100 | DEAD-box ATP-dependent RNA helicase 30 | Oryza sativa subsp japonica (Rice) | PR |
| Q0E2Z7 | Os02g0201900 | DEAD-box ATP-dependent RNA helicase 41 | Oryza sativa subsp japonica (Rice) | PR |
| Q9C718 | RH20 | DEAD-box ATP-dependent RNA helicase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8H136 | RH14 | DEAD-box ATP-dependent RNA helicase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4R3 | RH30 | DEAD-box ATP-dependent RNA helicase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SA27 | RH36 | DEAD-box ATP-dependent RNA helicase 36 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FVV4 | RH55 | DEAD-box ATP-dependent RNA helicase 55 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9CAI7 | TIF4A-3 | Eukaryotic initiation factor 4A-3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O80792 | RH33 | Putative DEAD-box ATP-dependent RNA helicase 33 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22907 | RH24 | DEAD-box ATP-dependent RNA helicase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LIH9 | RH51 | DEAD-box ATP-dependent RNA helicase 51 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8GY84 | RH10 | DEAD-box ATP-dependent RNA helicase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FFQ1 | RH31 | DEAD-box ATP-dependent RNA helicase 31 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SB89 | RH27 | DEAD-box ATP-dependent RNA helicase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q84TG1 | RH57 | DEAD-box ATP-dependent RNA helicase 57 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41377 | TIF4A-2 | Eukaryotic initiation factor 4A-2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41376 | EIF4A1 | Eukaryotic initiation factor 4A-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8H0U8 | RH42 | DEAD-box ATP-dependent RNA helicase 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVGASRTILS | LSLSSSLFTF | SKIPHVFPFL | RLHKPRFHHA | FRPLYSAAAT | TSSPTTETNV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TDPDQLKHTI | LLERLRLRHL | KESAKPPQQR | PSSVVGVEEE | SSIRKKSKKL | VENFQELGLS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EEVMGALQEL | NIEVPTEIQC | IGIPAVMERK | SVVLGSHTGS | GKTLAYLLPI | VQLMREDEAN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LGKKTKPRRP | RTVVLCPTRE | LSEQVYRVAK | SISHHARFRS | ILVSGGSRIR | PQEDSLNNAI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DMVVGTPGRI | LQHIEEGNMV | YGDIAYLVLD | EADTMFDRGF | GPEIRKFLAP | LNQRALKTND |
| 310 | 320 | 330 | 340 | 350 | 360 |
| QGFQTVLVTA | TMTMAVQKLV | DEEFQGIEHL | RTSTLHKKIA | NARHDFIKLS | GGEDKLEALL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QVLEPSLAKG | SKVMVFCNTL | NSSRAVDHYL | SENQISTVNY | HGEVPAEQRV | ENLKKFKDEE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GDCPTLVCTD | LAARGLDLDV | DHVVMFDFPK | NSIDYLHRTG | RTARMGAKGK | VTSLVSRKDQ |
| 490 | 500 | 510 | 520 | 530 | 540 |
| MLAARIEEAM | RNNESLESLT | TDNVRRDAAR | THITQEKGRS | VKQIREVSKQ | RNSRDKPSSS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SPPARSTGGK | TPVRKSSSSS | FSKPRKASSP | PEKSSKPKRK | ILKTVGSRSI | AARGKTGSDR |
| 610 | 620 | ||||
| RPGKKLSVVG | FRGKSSSARA | S |