Q4JF28
Gene name |
IRF3 |
Protein name |
Interferon regulatory factor 3 |
Names |
IRF-3 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:516979 |
EC number |
|
Protein Class |
|
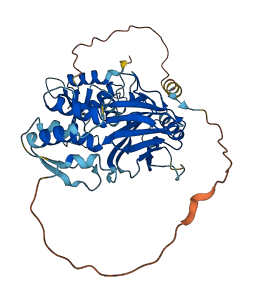
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
197-376 (IFN association domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Lin R et al. (1999) "Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains", Molecular and cellular biology, 19, 2465-74
- Shah M et al. (2015) "In silico mechanistic analysis of IRF3 inactivation and high-risk HPV E6 species-dependent drug response", Scientific reports, 5, 13446
- Liu S et al. (2015) "Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation", Science (New York, N.Y.), 347, aaa2630
Autoinhibited structure

Activated structure

1 structures for Q4JF28
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q4JF28-F1 | Predicted | AlphaFoldDB |
222 variants for Q4JF28
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs471186362 | 8 | I>M | No | Ensembl | |
| rs444527760 | 8 | I>R | No | Ensembl | |
| rs440734683 | 11 | W>C | No | Ensembl | |
| rs473654788 | 14 | S>F | No | Ensembl | |
| rs473654788 | 14 | S>Y | No | Ensembl | |
| rs469003634 | 17 | D>G | No | Ensembl | |
| rs435939956 | 17 | D>H | No | Ensembl | |
| rs435939956 | 17 | D>Y | No | Ensembl | |
| rs456999663 | 19 | G>A | No | Ensembl | |
| rs432110717 | 20 | E>A | No | Ensembl | |
| rs446564060 | 24 | V>G | No | Ensembl | |
| rs434551145 | 25 | A>S | No | Ensembl | |
| rs467476527 | 27 | L>P | No | Ensembl | |
| rs449114000 | 29 | E>A | No | Ensembl | |
| rs481205824 | 30 | S>R | No | Ensembl | |
| rs450973387 | 32 | T>A | No | Ensembl | |
| rs450973387 | 32 | T>P | No | Ensembl | |
| rs459058418 | 33 | R>P | No | Ensembl | |
| rs473606098 | 34 | F>L | No | Ensembl | |
| rs461623444 | 34 | F>S | No | Ensembl | |
| rs475440035 | 35 | R>P | No | Ensembl | |
| rs431967132 | 38 | W>L | No | Ensembl | |
| rs471426591 | 39 | K>Q | No | Ensembl | |
| rs452911707 | 40 | H>D | No | Ensembl | |
| rs452911707 | 40 | H>N | No | Ensembl | |
| rs434514346 | 41 | G>R | No | Ensembl | |
| rs437108652 | 42 | L>F | No | Ensembl | |
| rs449075509 | 42 | L>V | No | Ensembl | |
| rs469395541 | 43 | R>G | No | Ensembl | |
| rs450758925 | 44 | Q>P | No | Ensembl | |
| rs458946689 | 45 | D>E | No | Ensembl | |
| rs477445399 | 45 | D>G | No | Ensembl | |
| rs446997976 | 46 | A>G | No | Ensembl | |
| rs443134320 | 47 | Q>H | No | Ensembl | |
| rs461588662 | 47 | Q>P | No | Ensembl | |
| rs476064654 | 48 | Q>P | No | Ensembl | |
| rs476064654 | 48 | Q>R | No | Ensembl | |
| rs464065458 | 49 | E>G | No | Ensembl | |
| rs438355704 | 50 | D>G | No | Ensembl | |
| rs471419928 | 51 | F>V | No | Ensembl | |
| rs434477130 | 52 | G>C | No | Ensembl | |
| rs434477130 | 52 | G>R | No | Ensembl | |
| rs455418704 | 54 | F>L | No | Ensembl | |
| rs470023519 | 55 | Q>P | No | Ensembl | |
| rs463156259 | 61 | S>R | No | Ensembl | |
| rs471245348 | 63 | A>G | No | Ensembl | |
| rs452851366 | 65 | T>N | No | Ensembl | |
| rs433528728 | 67 | G>E | No | Ensembl | |
| rs473028191 | 72 | D>G | No | Ensembl | |
| rs454604597 | 89 | V>G | No | Ensembl | |
| rs1114509873 | 91 | R>C | No | Ensembl | |
| rs469349502 | 136 | E>* | No | Ensembl | |
| rs457278220 | 137 | D>E | No | Ensembl | |
| rs432366085 | 140 | E>G | No | Ensembl | |
| rs717915109 | 141 | K>N | No | Ensembl | |
| rs465428358 | 147 | D>A | No | Ensembl | |
| rs446997939 | 147 | D>E | No | Ensembl | |
| rs480047377 | 160 | S>F | No | Ensembl | |
| rs520313532 | 162 | K>N | No | Ensembl | |
| rs1116702921 | 163 | P>A | No | Ensembl | |
| rs1118251781 | 166 | F>L | No | Ensembl | |
| rs1118263930 | 168 | Q>L | No | Ensembl | |
| rs468039177 | 169 | S>G | No | Ensembl | |
| rs436119778 | 170 | P>H | No | Ensembl | |
| rs482532463 | 170 | P>T | No | Ensembl | |
| rs464025449 | 172 | S>A | No | Ensembl | |
| rs477627801 | 183 | L>I | No | Ensembl | |
| rs477627801 | 183 | L>V | No | Ensembl | |
| rs459260419 | 185 | E>D | No | Ensembl | |
| rs440900785 | 186 | N>T | No | Ensembl | |
| rs473996538 | 187 | P>A | No | Ensembl | |
| rs462000884 | 188 | L>M | No | Ensembl | |
| rs443398532 | 193 | A>G | No | Ensembl | |
| rs476512649 | 194 | N>D | No | Ensembl | |
| rs476512649 | 194 | N>H | No | Ensembl | |
| rs451643775 | 195 | E>A | No | Ensembl | |
| rs447192038 | 198 | W>G | No | Ensembl | |
| rs480298834 | 199 | E>A | No | Ensembl | |
| rs468276086 | 199 | E>D | No | Ensembl | |
| rs480298834 | 199 | E>G | No | Ensembl | |
| rs482808361 | 200 | F>C | No | Ensembl | |
| rs449738379 | 200 | F>I | No | Ensembl | |
| rs449738379 | 200 | F>L | No | Ensembl | |
| rs449738379 | 200 | F>V | No | Ensembl | |
| rs478866145 | 201 | E>G | No | Ensembl | |
| rs439431760 | 201 | E>K | No | Ensembl | |
| rs478866145 | 201 | E>V | No | Ensembl | |
| rs441167836 | 202 | V>A | No | Ensembl | |
| rs441167836 | 202 | V>E | No | Ensembl | |
| rs441167836 | 202 | V>G | No | Ensembl | |
| rs474236178 | 203 | T>S | No | Ensembl | |
| rs476603678 | 204 | A>G | No | Ensembl | |
| rs443733870 | 204 | A>S | No | Ensembl | |
| rs433356907 | 205 | F>L | No | Ensembl | |
| rs433356907 | 205 | F>V | No | Ensembl | |
| rs435161261 | 206 | Y>* | No | Ensembl | |
| rs454372581 | 206 | Y>C | No | Ensembl | |
| rs454372581 | 206 | Y>F | No | Ensembl | |
| rs466278644 | 206 | Y>H | No | Ensembl | |
| rs449764295 | 208 | G>C | No | Ensembl | |
| rs482669410 | 208 | G>V | No | Ensembl | |
| rs445828937 | 209 | C>F | No | Ensembl | |
| rs464266719 | 209 | C>S | No | Ensembl | |
| rs478827440 | 209 | C>W | No | Ensembl | |
| rs460431381 | 210 | Q>* | No | Ensembl | |
| rs441927538 | 211 | V>F | No | Ensembl | |
| rs480639800 | 211 | V>G | No | Ensembl | |
| rs441927538 | 211 | V>I | No | Ensembl | |
| rs462165297 | 212 | F>L | No | Ensembl | |
| rs443614023 | 213 | Q>R | No | Ensembl | |
| rs476468774 | 215 | T>P | No | Ensembl | |
| rs451704080 | 215 | T>S | No | Ensembl | |
| rs472643764 | 218 | C>W | No | Ensembl | |
| rs454403655 | 221 | G>R | No | Ensembl | |
| rs435936317 | 222 | L>Q | No | Ensembl | |
| rs435936317 | 222 | L>R | No | Ensembl | |
| rs467468053 | 225 | V>G | No | Ensembl | |
| rs468906341 | 225 | V>L | No | Ensembl | |
| rs437607696 | 226 | G>V | No | Ensembl | |
| rs445790334 | 230 | G>E | No | Ensembl | |
| rs433745996 | 231 | D>A | No | Ensembl | |
| rs433745996 | 231 | D>G | No | Ensembl | |
| rs466767684 | 233 | M>V | No | Ensembl | |
| rs448365043 | 234 | L>V | No | Ensembl | |
| rs462863846 | 235 | P>A | No | Ensembl | |
| rs482857189 | 237 | Q>* | No | Ensembl | |
| rs458065070 | 238 | P>A | No | Ensembl | |
| rs439706011 | 239 | I>K | No | Ensembl | |
| rs460654497 | 240 | R>P | No | Ensembl | |
| rs475273043 | 246 | T>A | No | Ensembl | |
| rs456841986 | 246 | T>R | No | Ensembl | |
| rs470616174 | 248 | L>M | No | Ensembl | |
| rs452216089 | 248 | L>P | No | Ensembl | |
| rs470616174 | 248 | L>V | No | Ensembl | |
| rs466686003 | 252 | S>R | No | Ensembl | |
| rs448144246 | 255 | D>A | No | Ensembl | |
| rs436203528 | 255 | D>E | No | Ensembl | |
| rs469222221 | 256 | Y>* | No | Ensembl | |
| rs450736164 | 257 | V>L | No | Ensembl | |
| rs482820067 | 258 | Q>H | No | Ensembl | |
| rs481828567 | 258 | Q>K | No | Ensembl | |
| rs458027910 | 259 | R>G | No | Ensembl | |
| rs446010134 | 260 | V>G | No | Ensembl | |
| rs479110882 | 263 | C>F | No | Ensembl | |
| rs475266305 | 269 | A>G | No | Ensembl | |
| rs442163966 | 269 | A>S | No | Ensembl | |
| rs470577158 | 271 | W>R | No | Ensembl | |
| rs452079358 | 273 | A>G | No | Ensembl | |
| rs439995841 | 274 | G>R | No | Ensembl | |
| rs473039196 | 275 | Q>E | No | Ensembl | |
| rs454632405 | 275 | Q>P | No | Ensembl | |
| rs454632405 | 275 | Q>R | No | Ensembl | |
| rs469247646 | 276 | W>G | No | Ensembl | |
| rs469247646 | 276 | W>R | No | Ensembl | |
| rs457072848 | 277 | L>I | No | Ensembl | |
| rs432206346 | 277 | L>R | No | Ensembl | |
| rs457072848 | 277 | L>V | No | Ensembl | |
| rs479077820 | 278 | C>* | No | Ensembl | |
| rs445972962 | 278 | C>F | No | Ensembl | |
| rs465108941 | 278 | C>G | No | Ensembl | |
| rs465108941 | 278 | C>R | No | Ensembl | |
| rs479077820 | 278 | C>W | No | Ensembl | |
| rs467010915 | 279 | A>D | No | Ensembl | |
| rs467010915 | 279 | A>G | No | Ensembl | |
| rs481664440 | 280 | Q>H | No | Ensembl | |
| rs448600767 | 280 | Q>P | No | Ensembl | |
| rs463197726 | 281 | R>G | No | Ensembl | |
| rs444648100 | 281 | R>K | No | Ensembl | |
| rs477654859 | 282 | L>P | No | Ensembl | |
| rs477654859 | 282 | L>R | No | Ensembl | |
| rs439961602 | 284 | H>D | No | Ensembl | |
| rs473060289 | 284 | H>P | No | Ensembl | |
| rs473060289 | 284 | H>R | No | Ensembl | |
| rs439961602 | 284 | H>Y | No | Ensembl | |
| rs442561317 | 285 | C>* | No | Ensembl | |
| rs454535799 | 285 | C>G | No | Ensembl | |
| rs454535799 | 285 | C>R | No | Ensembl | |
| rs475312542 | 286 | H>P | No | Ensembl | |
| rs432167356 | 287 | V>E | No | Ensembl | |
| rs432167356 | 287 | V>G | No | Ensembl | |
| rs456934709 | 287 | V>L | No | Ensembl | |
| rs433958787 | 288 | Y>C | No | Ensembl | |
| rs453116132 | 288 | Y>D | No | Ensembl | |
| rs467030755 | 290 | A>G | No | Ensembl | |
| rs448482052 | 291 | I>V | No | Ensembl | |
| rs481516234 | 292 | G>S | No | Ensembl | |
| rs477616487 | 293 | E>D | No | Ensembl | |
| rs451082041 | 293 | E>G | No | Ensembl | |
| rs459216077 | 294 | E>G | No | Ensembl | |
| rs479454197 | 295 | L>P | No | Ensembl | |
| rs440731785 | 295 | L>V | No | Ensembl | |
| rs475273909 | 299 | C>G | No | Ensembl | |
| rs438539972 | 302 | K>E | No | Ensembl | |
| rs471445289 | 302 | K>N | No | Ensembl | |
| rs434724189 | 307 | V>G | No | Ensembl | |
| rs474110693 | 308 | P>A | No | Ensembl | |
| rs436391641 | 317 | N>H | No | Ensembl | |
| rs469444856 | 318 | L>P | No | Ensembl | |
| rs432527569 | 321 | F>L | No | Ensembl | |
| rs465614401 | 324 | D>H | No | Ensembl | |
| rs447561004 | 327 | T>P | No | Ensembl | |
| rs480449228 | 330 | E>D | No | Ensembl | |
| rs462062017 | 331 | G>V | No | Ensembl | |
| rs442988817 | 334 | R>H | No | Ensembl | |
| rs442988817 | 334 | R>L | No | Ensembl | |
| rs439799855 | 339 | T>P | No | Ensembl | |
| rs439799855 | 339 | T>S | No | Ensembl | |
| rs472849202 | 341 | W>R | No | Ensembl | |
| rs454442452 | 342 | F>L | No | Ensembl | |
| rs474639860 | 343 | C>* | No | Ensembl | |
| rs435165852 | 343 | C>F | No | Ensembl | |
| rs474639860 | 343 | C>W | No | Ensembl | |
| rs437651038 | 346 | Q>E | No | Ensembl | |
| rs464266860 | 348 | W>S | No | Ensembl | |
| rs445758222 | 350 | Q>H | No | Ensembl | |
| rs480475867 | 351 | D>A | No | Ensembl | |
| rs466806425 | 357 | R>M | No | Ensembl | |
| rs447142067 | 372 | V>L | No | Ensembl | |
| rs457059922 | 388 | D>N | No | Ensembl | |
| rs1118272875 | 395 | D>E | No | Ensembl | |
| rs1118281959 | 395 | D>H | No | Ensembl | |
| rs1118279411 | 407 | C>Y | No | Ensembl |
No associated diseases with Q4JF28
Functions
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets transcribed by RNA polymerase II. |
| promoter-specific chromatin binding | Binding to a section of chromatin that is associated with gene promoter sequences of DNA. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
15 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to virus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a virus. |
| defense response to virus | Reactions triggered in response to the presence of a virus that act to protect the cell or organism. |
| immune system process | Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats. |
| lipopolysaccharide-mediated signaling pathway | The series of molecular signals initiated by the binding of a lipopolysaccharide (LPS) to a receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. Lipopolysaccharides are major components of the outer membrane of Gram-negative bacteria, making them prime targets for recognition by the immune system. |
| positive regulation of interferon-alpha production | Any process that activates or increases the frequency, rate, or extent of interferon-alpha production. |
| positive regulation of interferon-beta production | Any process that activates or increases the frequency, rate, or extent of interferon-beta production. |
| positive regulation of type I interferon production | Any process that activates or increases the frequency, rate, or extent of type I interferon production. Type I interferons include the interferon-alpha, beta, delta, episilon, zeta, kappa, tau, and omega gene families. |
| positive regulation of type I interferon-mediated signaling pathway | Any process that increases the rate, frequency or extent of a type I interferon-mediated signaling pathway. |
| programmed necrotic cell death | A necrotic cell death process that results from the activation of endogenous cellular processes, such as signaling involving death domain receptors or Toll-like receptors. |
| regulation of apoptotic process | Any process that modulates the occurrence or rate of cell death by apoptotic process. |
| regulation of inflammatory response | Any process that modulates the frequency, rate or extent of the inflammatory response, the immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| response to exogenous dsRNA | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an exogenous double-stranded RNA stimulus. |
| TRIF-dependent toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor where the TRIF adaptor mediates transduction of the signal. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
| type I interferon signaling pathway | The series of molecular signals initiated by type I interferon binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. Type I interferons include the interferon-alpha, beta, delta, episilon, zeta, kappa, tau, and omega gene families. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q58DJ0 | IRF5 | Interferon regulatory factor 5 | Bos taurus (Bovine) | SS |
| Q14653 | IRF3 | Interferon regulatory factor 3 | Homo sapiens (Human) | EV |
| P70671 | Irf3 | Interferon regulatory factor 3 | Mus musculus (Mouse) | SS |
| Q764M6 | IRF3 | Interferon regulatory factor 3 | Sus scrofa (Pig) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGTQKPRILP | WLISQLDRGE | LEGVAWLGES | RTRFRIPWKH | GLRQDAQQED | FGIFQAWAVA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SGAYTPGKDK | PDLPTWKRNF | RSALNRKEVL | RLAEDHSKDS | QDPHKIYEFV | NSGVRDIPEP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DTSQDNGRHN | TSDTQEDTLE | KLLSDMDLSP | GGPSNLTMAS | EKPPQFLQSP | DSDIPALCPN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SGLSENPLKQ | LLANEEDWEF | EVTAFYRGCQ | VFQQTVFCPG | GLRLVGSEAG | DRMLPGQPIR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LPDPATSLTD | KSVTDYVQRV | LSCLGGGLAL | WRAGQWLCAQ | RLGHCHVYWA | IGEELLPSCG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HKPDGEVPKD | REGGVFNLGP | FITDLITFIE | GSRRSPLYTL | WFCVGQSWPQ | DQPWIKRLVM |
| 370 | 380 | 390 | 400 | 410 | |
| VKVVPMCLRV | LVDIARQGGA | SSLENTVDLH | ISNSDPLSLT | PDQYMACLQD | LAEDMDF |