Q498D6
Gene name |
Fgfr4 |
Protein name |
Fibroblast growth factor receptor 4 |
Names |
|
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:25114 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
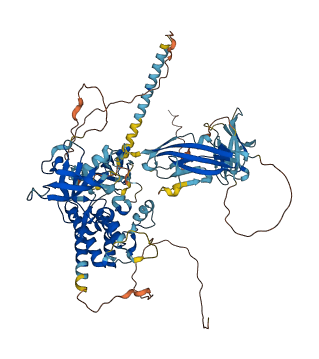
Autoinhibited structure

Activated structure

1 structures for Q498D6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q498D6-F1 | Predicted | AlphaFoldDB |
No variants for Q498D6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q498D6 | |||||
No associated diseases with Q498D6
14 regional properties for Q498D6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 465 - 753 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 466 - 741 | IPR001245 |
| domain | Immunoglobulin subtype 2 | 45 - 105 | IPR003598-1 |
| domain | Immunoglobulin subtype 2 | 159 - 227 | IPR003598-2 |
| domain | Immunoglobulin subtype 2 | 258 - 336 | IPR003598-3 |
| domain | Immunoglobulin subtype | 39 - 115 | IPR003599-1 |
| domain | Immunoglobulin subtype | 153 - 238 | IPR003599-2 |
| domain | Immunoglobulin subtype | 252 - 347 | IPR003599-3 |
| domain | Immunoglobulin-like domain | 148 - 236 | IPR007110-1 |
| domain | Immunoglobulin-like domain | 245 - 345 | IPR007110-2 |
| active_site | Tyrosine-protein kinase, active site | 606 - 618 | IPR008266 |
| domain | Immunoglobulin I-set | 157 - 235 | IPR013098 |
| binding_site | Protein kinase, ATP binding site | 471 - 501 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 465 - 741 | IPR020635 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| transport vesicle | Any of the vesicles of the constitutive secretory pathway, which carry cargo from the endoplasmic reticulum to the Golgi, between Golgi cisternae, from the Golgi to the ER (retrograde transport) or to destinations within or outside the cell. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| fibroblast growth factor binding | Binding to a fibroblast growth factor. |
| fibroblast growth factor receptor activity | Combining with a fibroblast growth factor receptor ligand and transmitting the signal across the plasma membrane to initiate a change in cell activity. |
| heparin binding | Binding to heparin, a member of a group of glycosaminoglycans found mainly as an intracellular component of mast cells and which consist predominantly of alternating alpha-(1->4)-linked D-galactose and N-acetyl-D-glucosamine-6-sulfate residues. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
27 GO annotations of biological process
| Name | Definition |
|---|---|
| alveolar secondary septum development | The progression of a secondary alveolar septum over time, from its formation to the mature structure. A secondary alveolar septum is a specialized epithelium that subdivides the initial saccule. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cholesterol homeostasis | Any process involved in the maintenance of an internal steady state of cholesterol within an organism or cell. |
| fibroblast growth factor receptor signaling pathway | The series of molecular signals generated as a consequence of a fibroblast growth factor receptor binding to one of its physiological ligands. |
| glucose homeostasis | Any process involved in the maintenance of an internal steady state of glucose within an organism or cell. |
| lung development | The process whose specific outcome is the progression of the lung over time, from its formation to the mature structure. In all air-breathing vertebrates the lungs are developed from the ventral wall of the oesophagus as a pouch which divides into two sacs. In amphibians and many reptiles the lungs retain very nearly this primitive sac-like character, but in the higher forms the connection with the esophagus becomes elongated into the windpipe and the inner walls of the sacs become more and more divided, until, in the mammals, the air spaces become minutely divided into tubes ending in small air cells, in the walls of which the blood circulates in a fine network of capillaries. In mammals the lungs are more or less divided into lobes, and each lung occupies a separate cavity in the thorax. |
| negative regulation of fibroblast growth factor production | Any process that decreases the rate, frequency or extent of the appearance of a fibroblast growth factor due to biosynthesis or secretion following a cellular stimulus, resulting in an increase in its intracellular or extracellular levels. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| organ induction | The interaction of two or more cells or tissues that causes them to change their fates and specify the development of an organ. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| phosphate ion homeostasis | Any process involved in the maintenance of an internal steady state of phosphate ions within an organism or cell. |
| positive regulation of catalytic activity | Any process that activates or increases the activity of an enzyme. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of DNA biosynthetic process | Any process that activates or increases the frequency, rate or extent of DNA biosynthetic process. |
| positive regulation of ERK1 and ERK2 cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of kinase activity | Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| positive regulation of parathyroid hormone secretion | Any process that activates or increases the frequency, rate or extent of parathyroid hormone secretion. |
| positive regulation of proteolysis | Any process that activates or increases the frequency, rate or extent of the hydrolysis of a peptide bond or bonds within a protein. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| regulation of bile acid biosynthetic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of bile acids. |
| regulation of extracellular matrix disassembly | Any process that modulates the rate, frequency or extent of extracellular matrix disassembly. Extracellular matrix disassembly is a process that results in the breakdown of the extracellular matrix. |
| regulation of lipid metabolic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving lipids. |
| regulation of phosphate transport | Any process that modulates the frequency, rate or extent of phosphate transport. Phosphate transport is the directed movement of phosphate into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of phosphorus metabolic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving phosphorus or compounds containing phosphorus. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| vitamin D3 metabolic process | The chemical reactions and pathways involving vitamin D3, (3S,5Z,7E)-9,10-secocholesta-5,7,10(19)-trien-3-ol. |
85 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P43481 | KIT | Mast/stem cell growth factor receptor Kit | Bos taurus (Bovine) | SS |
| Q06805 | TIE1 | Tyrosine-protein kinase receptor Tie-1 | Bos taurus (Bovine) | PR |
| Q06807 | TEK | Angiopoietin-1 receptor | Bos taurus (Bovine) | SS |
| Q28889 | KIT | Mast/stem cell growth factor receptor Kit | Felis catus (Cat) (Felis silvestris catus) | SS |
| P13369 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Felis catus (Cat) (Felis silvestris catus) | SS |
| P21804 | FGFR1 | Fibroblast growth factor receptor 1 | Gallus gallus (Chicken) | SS |
| Q9PUF6 | PDGFRA | Platelet-derived growth factor receptor alpha | Gallus gallus (Chicken) | SS |
| Q08156 | KIT | Mast/stem cell growth factor receptor Kit | Gallus gallus (Chicken) | SS |
| Q8QHL3 | FLT1 | Vascular endothelial growth factor receptor 1 | Gallus gallus (Chicken) | SS |
| P18460 | FGFR3 | Fibroblast growth factor receptor 3 | Gallus gallus (Chicken) | SS |
| P18461 | FGFR2 | Fibroblast growth factor receptor 2 | Gallus gallus (Chicken) | SS |
| Q07407 | htl | Fibroblast growth factor receptor homolog 1 | Drosophila melanogaster (Fruit fly) | PR |
| Q6J9G0 | STYK1 | Tyrosine-protein kinase STYK1 | Homo sapiens (Human) | PR |
| P36888 | FLT3 | Receptor-type tyrosine-protein kinase FLT3 | Homo sapiens (Human) | EV |
| P16234 | PDGFRA | Platelet-derived growth factor receptor alpha | Homo sapiens (Human) | EV |
| P09619 | PDGFRB | Platelet-derived growth factor receptor beta | Homo sapiens (Human) | EV |
| P35916 | FLT4 | Vascular endothelial growth factor receptor 3 | Homo sapiens (Human) | SS |
| P35968 | KDR | Vascular endothelial growth factor receptor 2 | Homo sapiens (Human) | EV |
| P17948 | FLT1 | Vascular endothelial growth factor receptor 1 | Homo sapiens (Human) | SS |
| P07333 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Homo sapiens (Human) | SS |
| P10721 | KIT | Mast/stem cell growth factor receptor Kit | Homo sapiens (Human) | EV |
| P07949 | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | Homo sapiens (Human) | EV |
| P35590 | TIE1 | Tyrosine-protein kinase receptor Tie-1 | Homo sapiens (Human) | PR |
| Q02763 | TEK | Angiopoietin-1 receptor | Homo sapiens (Human) | SS |
| P11362 | FGFR1 | Fibroblast growth factor receptor 1 | Homo sapiens (Human) | EV |
| P21802 | FGFR2 | Fibroblast growth factor receptor 2 | Homo sapiens (Human) | EV |
| P22607 | FGFR3 | Fibroblast growth factor receptor 3 | Homo sapiens (Human) | EV |
| P22455 | FGFR4 | Fibroblast growth factor receptor 4 | Homo sapiens (Human) | SS |
| P05532 | Kit | Mast/stem cell growth factor receptor Kit | Mus musculus (Mouse) | PR |
| Q91V87 | Fgfrl1 | Fibroblast growth factor receptor-like 1 | Mus musculus (Mouse) | PR |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| P09581 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Mus musculus (Mouse) | SS |
| P35969 | Flt1 | Vascular endothelial growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P35546 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Mus musculus (Mouse) | SS |
| Q06806 | Tie1 | Tyrosine-protein kinase receptor Tie-1 | Mus musculus (Mouse) | SS |
| Q00342 | Flt3 | Receptor-type tyrosine-protein kinase FLT3 | Mus musculus (Mouse) | SS |
| Q6J9G1 | Styk1 | Tyrosine-protein kinase STYK1 | Mus musculus (Mouse) | PR |
| Q61851 | Fgfr3 | Fibroblast growth factor receptor 3 | Mus musculus (Mouse) | PR |
| Q02858 | Tek | Angiopoietin-1 receptor | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| P21803 | Fgfr2 | Fibroblast growth factor receptor 2 | Mus musculus (Mouse) | SS |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| P16092 | Fgfr1 | Fibroblast growth factor receptor 1 | Mus musculus (Mouse) | SS |
| Q03142 | Fgfr4 | Fibroblast growth factor receptor 4 | Mus musculus (Mouse) | PR |
| Q2HWD6 | KIT | Mast/stem cell growth factor receptor Kit | Sus scrofa (Pig) | SS |
| Q7TQM3 | Fgfrl1 | Fibroblast growth factor receptor-like 1 | Rattus norvegicus (Rat) | PR |
| Q05030 | Pdgfrb | Platelet-derived growth factor receptor beta | Rattus norvegicus (Rat) | SS |
| G3V9H8 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Rattus norvegicus (Rat) | SS |
| Q62838 | Musk | Muscle, skeletal receptor tyrosine protein kinase | Rattus norvegicus (Rat) | SS |
| Q63604 | Ntrk2 | BDNF/NT-3 growth factors receptor | Rattus norvegicus (Rat) | SS |
| P35739 | Ntrk1 | High affinity nerve growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q03351 | Ntrk3 | NT-3 growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q91ZT1 | Flt4 | Vascular endothelial growth factor receptor 3 | Rattus norvegicus (Rat) | SS |
| O08775 | Kdr | Vascular endothelial growth factor receptor 2 | Rattus norvegicus (Rat) | SS |
| Q64716 | Insrr | Insulin receptor-related protein | Rattus norvegicus (Rat) | SS |
| P24062 | Igf1r | Insulin-like growth factor 1 receptor | Rattus norvegicus (Rat) | SS |
| P57097 | Mertk | Tyrosine-protein kinase Mer | Rattus norvegicus (Rat) | SS |
| P97523 | Met | Hepatocyte growth factor receptor | Rattus norvegicus (Rat) | PR |
| Q62956 | Erbb4 | Receptor tyrosine-protein kinase erbB-4 | Rattus norvegicus (Rat) | SS |
| Q62799 | Erbb3 | Receptor tyrosine-protein kinase erbB-3 | Rattus norvegicus (Rat) | SS |
| P20786 | Pdgfra | Platelet-derived growth factor receptor alpha | Rattus norvegicus (Rat) | SS |
| P53767 | Flt1 | Vascular endothelial growth factor receptor 1 | Rattus norvegicus (Rat) | PR |
| Q04589 | Fgfr1 | Fibroblast growth factor receptor 1 | Rattus norvegicus (Rat) | SS |
| Q63474 | Ddr1 | Epithelial discoidin domain-containing receptor 1 | Rattus norvegicus (Rat) | SS |
| P15127 | Insr | Insulin receptor | Rattus norvegicus (Rat) | SS |
| P06494 | Erbb2 | Receptor tyrosine-protein kinase erbB-2 | Rattus norvegicus (Rat) | SS |
| Q00495 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Rattus norvegicus (Rat) | PR |
| Q17833 | old-1 | Tyrosine-protein kinase receptor old-1 | Caenorhabditis elegans | PR |
| Q19238 | F09A5.2 | Putative tyrosine-protein kinase F09A5.2 | Caenorhabditis elegans | SS |
| P34892 | kin-16 | Receptor-like tyrosine-protein kinase kin-16 | Caenorhabditis elegans | PR |
| G5ED65 | ver-1 | Protein ver-1 | Caenorhabditis elegans | PR |
| Q10656 | egl-15 | Myoblast growth factor receptor egl-15 | Caenorhabditis elegans | PR |
| Q9S9M2 | WAKL4 | Wall-associated receptor kinase-like 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7X8C5 | WAKL2 | Wall-associated receptor kinase-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8AXB3 | kdrl | Vascular endothelial growth factor receptor kdr-like | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5GIT4 | kdr | Vascular endothelial growth factor receptor 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73791 | tek | Angiopoietin-1 receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q8JG38 | fgfr2 | Fibroblast growth factor receptor 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9I8N6 | csf1r | Macrophage colony-stimulating factor 1 receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9DE49 | pdgfra | Platelet-derived growth factor receptor alpha | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q8JFR5 | kita | Mast/stem cell growth factor receptor kita | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5MD89 | flt4 | Vascular endothelial growth factor receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q90Z00 | fgfr1a | Fibroblast growth factor receptor 1-A | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q90413 | fgfr4 | Fibroblast growth factor receptor 4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MWLLLALLSI | FQETPAFSLE | ASEEMEQEPC | PAPISEQQEQ | VLTVALGQPV | RLCCGRTERG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RHWYKEGSRL | ASAGRVRGWR | GRLEIASFLP | EDAGRYLCLA | RGSMTVVHNL | TLIMDDSLPS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| INNEDPKTLS | SSSSGHSYLQ | QAPYWTHPQR | MEKKLHAVPA | GNTVKFRCPA | AGNPMPTIHW |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LKNGQAFHGE | NRIGGIRLRH | QHWSLVMESV | VPSDRGTYTC | LVENSLGSIR | YSYLLDVLER |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SPHRPILQAG | LPANTTAVVG | SNVELLCKVY | SDAQPHIQWL | KHIVINGSSF | GADGFPYVQV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LKTTDINSSE | VEVLYLRNVS | AEDAGEYTCL | AGNSIGLSYQ | SAWLTVLPAE | EEDLAWTTAT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SEARYTDIIL | YVSGSLALVL | LLLLAGVYHR | QAIHGHHSRQ | PVTVQKLSRF | PLARQFSLES |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RSSGKSSLSL | VRGVRLSSSG | PPLLTGLVSL | DLPLDPLWEF | PRDRLVLGKP | LGEGCFGQVV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RAEALGMDSS | RPDQTSTVAV | KMLKDNASDK | DLADLISEME | MMKLIGRHKN | IINLLGVCTQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EGPLYVIVEY | AAKGNLREFL | RARRPPGPDL | SPDGPRSSEG | PLSFPALVSC | AYQVARGMQY |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LESRKCIHRD | LAARNVLVTE | DDVMKIADFG | LARGVHHIDY | YKKTSNGRLP | VKWMAPEALF |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DRVYTHQSDV | WSFGILLWEI | FTLGGSPYPG | IPVEELFSLL | REGHRMERPP | NCPSELYGLM |
| 730 | 740 | 750 | 760 | 770 | 780 |
| RECWHAAPSQ | RPTFKQLVEA | LDKVLLAVSE | EYLDLRLTFG | PYSPNNGDAS | STCSSSDSVF |
| 790 | |||||
| SHDPLPLEPS | PFPFPEAQTT |